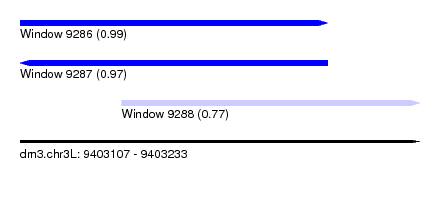
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,403,107 – 9,403,233 |
| Length | 126 |
| Max. P | 0.993266 |

| Location | 9,403,107 – 9,403,204 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 78.14 |
| Shannon entropy | 0.42298 |
| G+C content | 0.43687 |
| Mean single sequence MFE | -27.39 |
| Consensus MFE | -21.73 |
| Energy contribution | -23.10 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.993266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
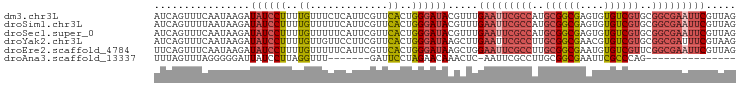
>dm3.chr3L 9403107 97 + 24543557 AUCAGUUUCAAUAAGAUAUCCUUUUGUUUCUCAUUCGUUCACUGGGAUACGUUUGAAUUCGCCAUGCGGCGAGUGUGUCGUGCGGCGAAUUCGUUAG ................((((((..((...(......)..))..)))))).....(((((((((..((((((....))))))..)))))))))..... ( -28.80, z-score = -2.21, R) >droSim1.chr3L 8792610 97 + 22553184 AUCAGUUUUAAUAAGAUAUCCUUUUGUUUUUCAUUCGUUCACUGGGAUACGUUUGAAUUCGCCAUGCGGCGAGUGUGUCGUGCGGCGAAUUCGUUAG ................((((((..((.............))..)))))).....(((((((((..((((((....))))))..)))))))))..... ( -28.42, z-score = -2.36, R) >droSec1.super_0 1648261 97 + 21120651 AUCAGUUUCAAUAAGAUAUCCUUUUGUUUUUCAUUCGUUCACUGGGAUACGUUUGAAUUCGCCAUGCGGCGAGUGUGUCGUGCGGCGAAUUCGUUAG ................((((((..((.............))..)))))).....(((((((((..((((((....))))))..)))))))))..... ( -28.42, z-score = -2.24, R) >droYak2.chr3L 21680061 97 - 24197627 AUCAGUUUCAAUAAGAUAUCCUUUUGUUGUUCCUUCGUUCACUGGGAUAAGCUUGAAUUCGCCUUGCGGCGAACGUGUCGUGCGGCGAUUUCGUAAG ...(((((((((((((.....)))))))).((((.........)))).))))).(((.(((((.(((((((....))))))).))))).)))..... ( -26.20, z-score = -1.18, R) >droEre2.scaffold_4784 9398086 97 + 25762168 UUCAGUUUCAAUAAGAUAUCCUUUUGUUUUUCAUUCGUUCACUGGGAUAAGCUGGAAUUCGCCUUGCGGCGAAUGUGUCGUUCGGCGAAUUCGUUAG ..(((((((.....))((((((..((.............))..)))))))))))(((((((((..((((((....))))))..)))))))))..... ( -30.92, z-score = -3.41, R) >droAna3.scaffold_13337 16989630 74 + 23293914 UUUAGUUUAGGGGGAUUAUCCUUAGGUUU-------GAUUCCUAGAACAAACUC-AAUUCGCCUUGCGGCGAAUUCGCCCAG--------------- .........(((.((.........(((((-------(.(((...))))))))).-.(((((((....))))))))).)))..--------------- ( -21.60, z-score = -1.94, R) >consensus AUCAGUUUCAAUAAGAUAUCCUUUUGUUUUUCAUUCGUUCACUGGGAUAAGCUUGAAUUCGCCAUGCGGCGAAUGUGUCGUGCGGCGAAUUCGUUAG ................((((((..((.............))..)))))).....(((((((((..((((((....))))))..)))))))))..... (-21.73 = -23.10 + 1.38)
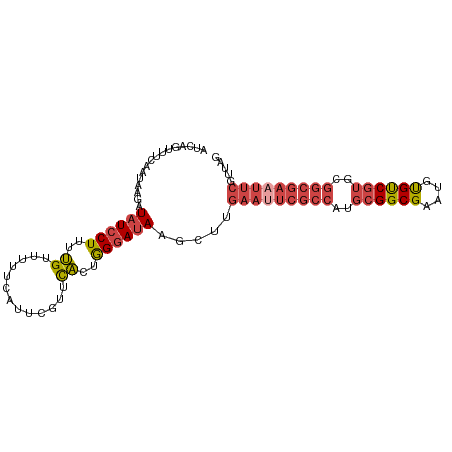
| Location | 9,403,107 – 9,403,204 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 78.14 |
| Shannon entropy | 0.42298 |
| G+C content | 0.43687 |
| Mean single sequence MFE | -22.04 |
| Consensus MFE | -12.69 |
| Energy contribution | -12.92 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.970526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9403107 97 - 24543557 CUAACGAAUUCGCCGCACGACACACUCGCCGCAUGGCGAAUUCAAACGUAUCCCAGUGAACGAAUGAGAAACAAAAGGAUAUCUUAUUGAAACUGAU .....(((((((((((.(((.....)))..))..)))))))))..........((((...(.(((((((............))))))))..)))).. ( -23.70, z-score = -2.68, R) >droSim1.chr3L 8792610 97 - 22553184 CUAACGAAUUCGCCGCACGACACACUCGCCGCAUGGCGAAUUCAAACGUAUCCCAGUGAACGAAUGAAAAACAAAAGGAUAUCUUAUUAAAACUGAU .....(((((((((((.(((.....)))..))..)))))))))....((((((...((.............))...))))))............... ( -21.32, z-score = -2.50, R) >droSec1.super_0 1648261 97 - 21120651 CUAACGAAUUCGCCGCACGACACACUCGCCGCAUGGCGAAUUCAAACGUAUCCCAGUGAACGAAUGAAAAACAAAAGGAUAUCUUAUUGAAACUGAU .....(((((((((((.(((.....)))..))..)))))))))....((((((...((.............))...))))))............... ( -21.32, z-score = -2.16, R) >droYak2.chr3L 21680061 97 + 24197627 CUUACGAAAUCGCCGCACGACACGUUCGCCGCAAGGCGAAUUCAAGCUUAUCCCAGUGAACGAAGGAACAACAAAAGGAUAUCUUAUUGAAACUGAU ...(((...(((.....)))..)))(((((....))))).(((((...(((((...((..(....)..))......))))).....)))))...... ( -21.00, z-score = -1.08, R) >droEre2.scaffold_4784 9398086 97 - 25762168 CUAACGAAUUCGCCGAACGACACAUUCGCCGCAAGGCGAAUUCCAGCUUAUCCCAGUGAACGAAUGAAAAACAAAAGGAUAUCUUAUUGAAACUGAA .....(((((((((...((.(......).))...)))))))))(((..(((((...((.............))...)))))((.....))..))).. ( -19.62, z-score = -1.68, R) >droAna3.scaffold_13337 16989630 74 - 23293914 ---------------CUGGGCGAAUUCGCCGCAAGGCGAAUU-GAGUUUGUUCUAGGAAUC-------AAACCUAAGGAUAAUCCCCCUAAACUAAA ---------------..(((.(((((((((....))))))))-....((((((((((....-------...))).)))))))..))))......... ( -25.30, z-score = -3.59, R) >consensus CUAACGAAUUCGCCGCACGACACACUCGCCGCAAGGCGAAUUCAAACGUAUCCCAGUGAACGAAUGAAAAACAAAAGGAUAUCUUAUUGAAACUGAU .........................(((((....))))).(((((..((((((...((.............))...))))))....)))))...... (-12.69 = -12.92 + 0.23)

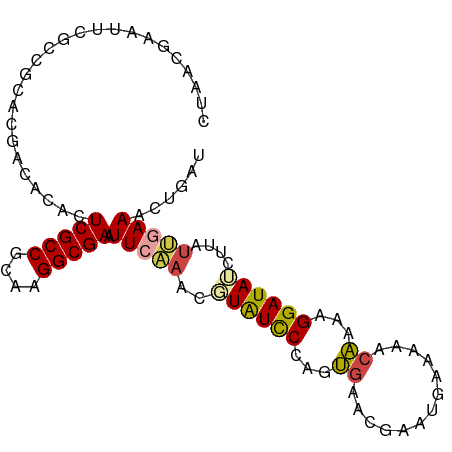
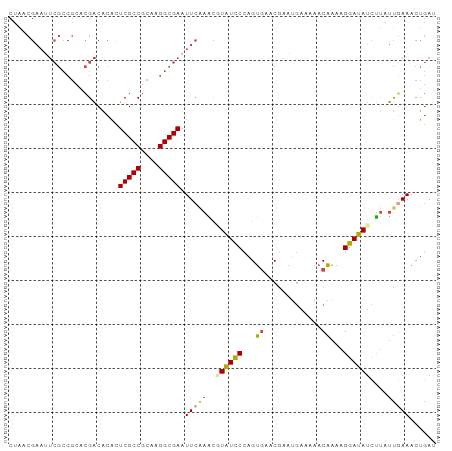
| Location | 9,403,139 – 9,403,233 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 84.68 |
| Shannon entropy | 0.29584 |
| G+C content | 0.49781 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -20.19 |
| Energy contribution | -21.47 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.768899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9403139 94 + 24543557 AUUCGUUCACUGGGAUACGUUUGAAUUCGCCAUGCGGCGAGUGUGUCGUGCGGCGAAUUCGUUAGCCGUGUUUUUCGUCCUCUUUGUUCUACCA ....(..(...((((((((..((((((((((..((((((....))))))..)))))))))).....))))))))..)..).............. ( -27.90, z-score = -1.24, R) >droSim1.chr3L 8792642 94 + 22553184 AUUCGUUCACUGGGAUACGUUUGAAUUCGCCAUGCGGCGAGUGUGUCGUGCGGCGAAUUCGUUAGCCGUGUUUUUCGUCCUCUUUGUUCUACCA ....(..(...((((((((..((((((((((..((((((....))))))..)))))))))).....))))))))..)..).............. ( -27.90, z-score = -1.24, R) >droSec1.super_0 1648293 94 + 21120651 AUUCGUUCACUGGGAUACGUUUGAAUUCGCCAUGCGGCGAGUGUGUCGUGCGGCGAAUUCGUUAGCCGUGUUUUUCGUCCUCUUUGUUCUACCA ....(..(...((((((((..((((((((((..((((((....))))))..)))))))))).....))))))))..)..).............. ( -27.90, z-score = -1.24, R) >droYak2.chr3L 21680093 94 - 24197627 CUUCGUUCACUGGGAUAAGCUUGAAUUCGCCUUGCGGCGAACGUGUCGUGCGGCGAUUUCGUAAGCCGUGUUUUUCGUCCUCUUUGUUCUACCA ..........(((((((((((((((.(((((.(((((((....))))))).))))).)))..))))((.......))......))))))))... ( -25.40, z-score = -0.44, R) >droEre2.scaffold_4784 9398118 94 + 25762168 AUUCGUUCACUGGGAUAAGCUGGAAUUCGCCUUGCGGCGAAUGUGUCGUUCGGCGAAUUCGUUAGCCGUGUUUUUCGUCCUCUUUGUUCUACCA ..........(((((((((((((((((((((..((((((....))))))..)))))))))..))))((.......))......))))))))... ( -28.90, z-score = -1.92, R) >droAna3.scaffold_13337 16989659 76 + 23293914 ----GAUUCCUAGAACAAACUC-AAUUCGCCUUGCGGCGAAU---UCGCCCAGC-----CGU-----GUUUUCUUCGUCCUCUUUGUUCUACCA ----......(((((((((...-((((((((....)))))))---)(((.....-----.))-----)..............)))))))))... ( -18.60, z-score = -3.39, R) >consensus AUUCGUUCACUGGGAUAAGCUUGAAUUCGCCAUGCGGCGAAUGUGUCGUGCGGCGAAUUCGUUAGCCGUGUUUUUCGUCCUCUUUGUUCUACCA ..........(((((((.....(((((((((..((((((....))))))..)))))))))......((.......)).......)))))))... (-20.19 = -21.47 + 1.28)

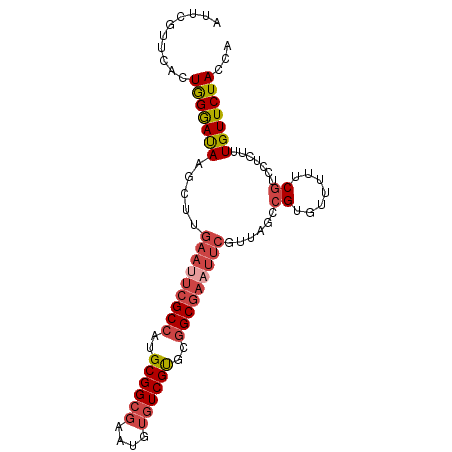
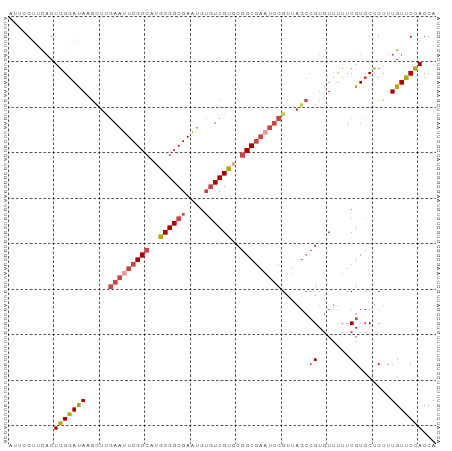
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:13:42 2011