| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,342,398 – 9,342,488 |
| Length | 90 |
| Max. P | 0.945399 |

| Location | 9,342,398 – 9,342,488 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 71.03 |
| Shannon entropy | 0.51777 |
| G+C content | 0.46572 |
| Mean single sequence MFE | -26.25 |
| Consensus MFE | -13.86 |
| Energy contribution | -14.17 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.945399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
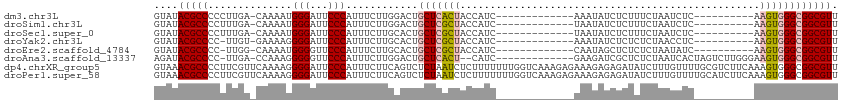
>dm3.chr3L 9342398 90 + 24543557 GUAUACGCCCCCUUGA-CAAAAUGGGAUUCCCAUUUCUUGGACUGCUCACUACCAUC-------------AAAUAUCUCUUUCUAAUCUC----------AAGUGGGCGGCGUU ....(((((.((..((-...((((((...))))))))..))...(((((((......-------------....................----------.)))))))))))). ( -23.93, z-score = -2.68, R) >droSim1.chr3L 8735680 90 + 22553184 GUAUACGCCCCUUUGA-CAAAAUGGGAUUCCCAUUUCUUGGACUGCUCGCUACCAUC-------------UAAUAUCUCUUUCUAAUCUC----------AAGUGGGCGGCGUU ....(((((((...((-...((((((...))))))))..))...(((((((......-------------....................----------.)))))))))))). ( -22.23, z-score = -1.91, R) >droSec1.super_0 1591936 90 + 21120651 GUAUACGCCCCUUUGA-CAAAAUGGGAUUCCCAUUUCUUGCACUGCUCGCUACCAUC-------------UAAUAUCUCUUUCUAAUCUC----------AAGUGGGCGGCGUU ....(((((....((.-(((((((((...))))))..)))))..(((((((......-------------....................----------.)))))))))))). ( -21.33, z-score = -2.16, R) >droYak2.chr3L 21618330 89 - 24197627 GUAUACGCCCC-UUGU-GAAAAGGGGAUUCCCAUUUCUUGCACUGCUCGCUACCAUC-------------AAAUAUCUCUCUCUAACCUC----------AAGUGGGCGGCGUU .......((((-((..-...)))))).............((.(((((((((......-------------....................----------.))))))))).)). ( -22.13, z-score = -1.30, R) >droEre2.scaffold_4784 9342043 89 + 25762168 GUAUACGCCCC-UUGG-CAAAAUGGGGUUCCCAUUUCUUGCACUGCUCGCUACCAUC-------------CAAUAGCUCUCUCUAAUAUC----------AAGUGGGCGGCGUU .....((((((-((((-(((((((((...))))))..)))).......((((.....-------------...))))............)----------))).)))))..... ( -27.70, z-score = -2.41, R) >droAna3.scaffold_13337 16932761 97 + 23293914 AGAUACGCCCC-UUGA-CCAAAGGGGGUUCCCAUUUCUUGGACUGCUCACU--CAUC-------------GAAGAUCGCUCUCUAAUCACUAGUCUUGGGAAGUGGGCGGCGUU ......(((((-((..-....)))))))..(((.....))).(((((((((--..((-------------.(((((................))))).)).))))))))).... ( -29.99, z-score = -0.47, R) >dp4.chrXR_group5 555700 114 - 740492 GUAAACGCCCCUUCGUUCAAAAGGGGAUUCCCAUUUCUUCAGUCUCUAAUCUCUUUUUUUGGUCAAAGAGAAAGAGAGAUAUCUUUGUUUUGCGUCUUCAAAGUGGGCGGCGUU ...(((((((((.(((.((((.(((....))).........((((((..(((((((........)))))))...))))))...))))....)))..........))).)))))) ( -31.70, z-score = -1.75, R) >droPer1.super_58 231443 114 + 410386 GUAAACGCCCCUUCGUUCAAAAGGGGAUUCCCAUUUCUUCAGUCUCUAAUCUCUUUUUUUGGUCAAAGAGAAAGAGAGAUAUCUUUGUUUUGCAUCUUCAAAGUGGGCGGCGUU ...(((((((((..........(((....))).........((((((..(((((((........)))))))...))))))..(((((...........))))).))).)))))) ( -31.00, z-score = -1.71, R) >consensus GUAUACGCCCCUUUGA_CAAAAGGGGAUUCCCAUUUCUUGCACUGCUCACUACCAUC_____________AAAUAUCUCUCUCUAAUCUC__________AAGUGGGCGGCGUU ....(((((..............(((...)))............(((((((..................................................)))))))))))). (-13.86 = -14.17 + 0.31)
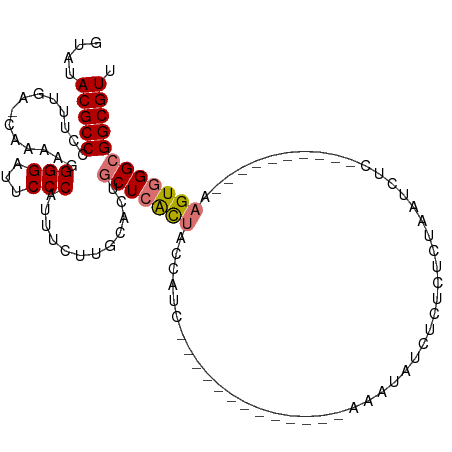
| Location | 9,342,398 – 9,342,488 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 71.03 |
| Shannon entropy | 0.51777 |
| G+C content | 0.46572 |
| Mean single sequence MFE | -27.49 |
| Consensus MFE | -11.31 |
| Energy contribution | -10.62 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.874191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9342398 90 - 24543557 AACGCCGCCCACUU----------GAGAUUAGAAAGAGAUAUUU-------------GAUGGUAGUGAGCAGUCCAAGAAAUGGGAAUCCCAUUUUG-UCAAGGGGGCGUAUAC .(((((.(((.(((----------(.((((.(.......((((.-------------...)))).....)))))))))(((((((...)))))))..-....)))))))).... ( -26.20, z-score = -2.28, R) >droSim1.chr3L 8735680 90 - 22553184 AACGCCGCCCACUU----------GAGAUUAGAAAGAGAUAUUA-------------GAUGGUAGCGAGCAGUCCAAGAAAUGGGAAUCCCAUUUUG-UCAAAGGGGCGUAUAC .....(((((..((----------((..................-------------..(((..((.....))))).((((((((...)))))))).-))))..)))))..... ( -23.20, z-score = -1.81, R) >droSec1.super_0 1591936 90 - 21120651 AACGCCGCCCACUU----------GAGAUUAGAAAGAGAUAUUA-------------GAUGGUAGCGAGCAGUGCAAGAAAUGGGAAUCCCAUUUUG-UCAAAGGGGCGUAUAC .....(((((..((----------((..................-------------.......(((.....)))..((((((((...)))))))).-))))..)))))..... ( -22.80, z-score = -1.71, R) >droYak2.chr3L 21618330 89 + 24197627 AACGCCGCCCACUU----------GAGGUUAGAGAGAGAUAUUU-------------GAUGGUAGCGAGCAGUGCAAGAAAUGGGAAUCCCCUUUUC-ACAA-GGGGCGUAUAC .....(((((.(((----------(..((((((........)))-------------)))....(((.....)))..((((.(((....))).))))-.)))-))))))..... ( -25.10, z-score = -1.42, R) >droEre2.scaffold_4784 9342043 89 - 25762168 AACGCCGCCCACUU----------GAUAUUAGAGAGAGCUAUUG-------------GAUGGUAGCGAGCAGUGCAAGAAAUGGGAACCCCAUUUUG-CCAA-GGGGCGUAUAC .....(((((.(((----------(........(...((((((.-------------...))))))...)...((((..((((((...)))))))))-))))-))))))..... ( -29.00, z-score = -2.44, R) >droAna3.scaffold_13337 16932761 97 - 23293914 AACGCCGCCCACUUCCCAAGACUAGUGAUUAGAGAGCGAUCUUC-------------GAUG--AGUGAGCAGUCCAAGAAAUGGGAACCCCCUUUGG-UCAA-GGGGCGUAUCU .(((((((.(((((..(((((...((.........))..)))).-------------)..)--)))).)).(.(((((....(((....))))))))-.)..-..))))).... ( -26.80, z-score = -0.36, R) >dp4.chrXR_group5 555700 114 + 740492 AACGCCGCCCACUUUGAAGACGCAAAACAAAGAUAUCUCUCUUUCUCUUUGACCAAAAAAAGAGAUUAGAGACUGAAGAAAUGGGAAUCCCCUUUUGAACGAAGGGGCGUUUAC ((((((.(((((((((...........))))).....(((((.(((((((........)))))))..))))).........)))).....(((((.....)))))))))))... ( -33.40, z-score = -3.39, R) >droPer1.super_58 231443 114 - 410386 AACGCCGCCCACUUUGAAGAUGCAAAACAAAGAUAUCUCUCUUUCUCUUUGACCAAAAAAAGAGAUUAGAGACUGAAGAAAUGGGAAUCCCCUUUUGAACGAAGGGGCGUUUAC ((((((.(((((((((...........))))).....(((((.(((((((........)))))))..))))).........)))).....(((((.....)))))))))))... ( -33.40, z-score = -3.42, R) >consensus AACGCCGCCCACUU__________GAGAUUAGAAAGAGAUAUUU_____________GAUGGUAGCGAGCAGUCCAAGAAAUGGGAAUCCCAUUUUG_UCAAAGGGGCGUAUAC .(((((.((..................................................((....))..........((((.(((...))).)))).......))))))).... (-11.31 = -10.62 + -0.69)

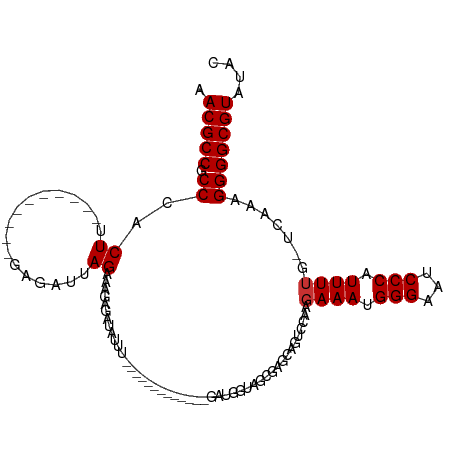
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:13:35 2011