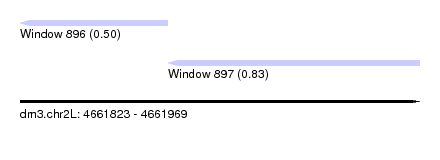
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,661,823 – 4,661,969 |
| Length | 146 |
| Max. P | 0.834242 |

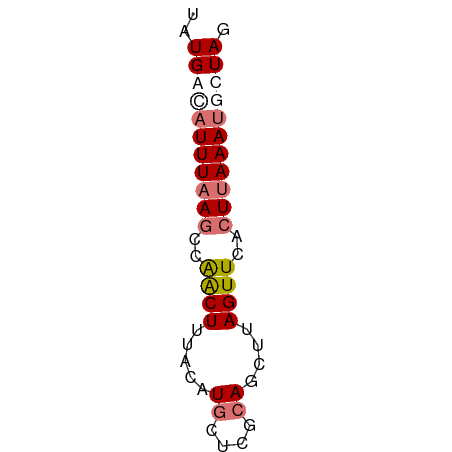
| Location | 4,661,823 – 4,661,877 |
|---|---|
| Length | 54 |
| Sequences | 4 |
| Columns | 54 |
| Reading direction | reverse |
| Mean pairwise identity | 81.79 |
| Shannon entropy | 0.29943 |
| G+C content | 0.34862 |
| Mean single sequence MFE | -9.20 |
| Consensus MFE | -6.86 |
| Energy contribution | -8.42 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2L 4661823 54 - 23011544 UAUGAUAUUUAAUCCAACUUUACAUGCUCGCAGUUUAGUUCACUUAAAUGCUAG .....(((((((...((((..((.((....))))..))))...))))))).... ( -6.50, z-score = -0.84, R) >droSim1.chr2L 4533809 54 - 22036055 UAUGGCAUUUAAGCCGACUUUACAUGCUCGCAGCUUAGUUCACUUAAAUGCUAG ..(((((((((((..((((.....((....))....))))..))))))))))). ( -16.50, z-score = -3.39, R) >droSec1.super_5 2746127 54 - 5866729 UAUGGCAUUUUAGCUAACUUUACAUGCUCGCAGCUUAGUUCACUUAAAUCCUAG ...((.((((.((..((((.....((....))....))))..)).))))))... ( -6.30, z-score = 0.25, R) >droYak2.chr2L 4674754 53 - 22324452 AAUGACAUUUAAGCCAGCUUUACAUGUACGGAG-AUAGUUGACUUAAAAUAUAU .......((((((.(((((...(......)...-..))))).))))))...... ( -7.50, z-score = -1.27, R) >consensus UAUGACAUUUAAGCCAACUUUACAUGCUCGCAGCUUAGUUCACUUAAAUGCUAG ..(((((((((((..((((.....((....))....))))..))))))))))). ( -6.86 = -8.42 + 1.56)


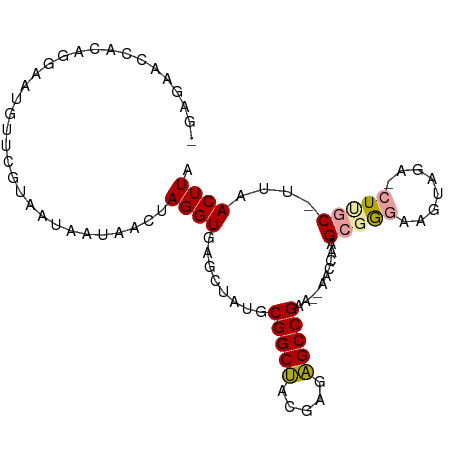
| Location | 4,661,877 – 4,661,969 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 66.43 |
| Shannon entropy | 0.47644 |
| G+C content | 0.48246 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -10.14 |
| Energy contribution | -10.37 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4661877 92 - 23011544 -GAGAACCACA-GCAUGUUGGUUGUCAUAUCAAGGUGAGGUAUGCGGCAGCGAGUGCCGAG--AACCCUGAUGCAAGUGGAACUUGCGUUAACUUG -((.(((((..-......))))).))....((((....(((...(((((.....)))))..--.))).(((((((((.....))))))))).)))) ( -31.00, z-score = -1.96, R) >droSec1.super_2164 216 93 - 3633 -GAGAAUCACAGGAAUGCUUGGAAUAUUGACUAGGUGAGCUGUGCGGCUACGAGAGCCGGAGUAGAAAAGCGAGGAGUAGA-CUCGC-UUAACUUA -......(((((...(((((((........)))))))..)))))(((((.....)))))((((....(((((((.......-)))))-)).)))). ( -28.50, z-score = -2.29, R) >droEre2.scaffold_4845 20419826 95 + 22589142 CGAGAACAUCCGGAAUGUUCUUCAUACUUACUAGGUGAGCUAUGCGGCUACGAGAGCCGCAUCAACCAAGCGGGAAAUAUAUCUUGC-UUAACUUA .((((((((.....))))))))...........(((.....((((((((.....))))))))..)))((((((((......))))))-))...... ( -31.00, z-score = -3.51, R) >consensus _GAGAACCACAGGAAUGUUCGUAAUAAUAACUAGGUGAGCUAUGCGGCUACGAGAGCCGAA__AACCAAGCGGGAAGUAGA_CUUGC_UUAACUUA ................................((((........(((((.....)))))..........(((((........)))))....)))). (-10.14 = -10.37 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:17:09 2011