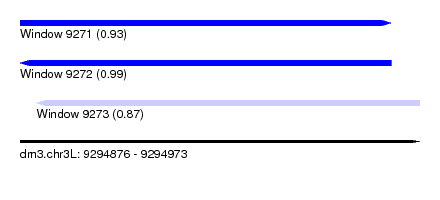
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,294,876 – 9,294,973 |
| Length | 97 |
| Max. P | 0.993186 |

| Location | 9,294,876 – 9,294,966 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 91.10 |
| Shannon entropy | 0.18343 |
| G+C content | 0.33881 |
| Mean single sequence MFE | -19.65 |
| Consensus MFE | -13.95 |
| Energy contribution | -13.92 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.925799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
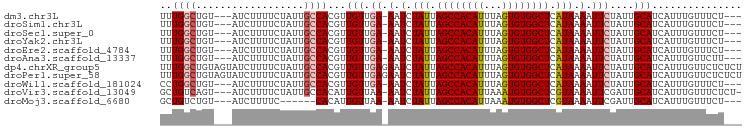
>dm3.chr3L 9294876 90 + 24543557 UUUGGCUGU---AUCUUUUCUAUUGCCACGUUGUUGA-AAUCUAUUAGCCACAUUUAGUGUGGCUCAUAAAAUUCUAUUGCAUCAUUUGUUUCU--- ..((((.((---(.......))).)))).((.((.((-(.(.(((.((((((((...)))))))).))).).))).)).)).............--- ( -19.10, z-score = -2.47, R) >droSim1.chr3L 8684490 90 + 22553184 UUUGGCUGU---AUCUUUUCUAUUGCCACGUUGUUGA-AAUCUAUUAGCCACAUUUAGUGUGGCUCAUAAAAUUCUAUUGCAUCAUUUGUUUCU--- ..((((.((---(.......))).)))).((.((.((-(.(.(((.((((((((...)))))))).))).).))).)).)).............--- ( -19.10, z-score = -2.47, R) >droSec1.super_0 1543806 90 + 21120651 UUUGGCUGU---AUCUUUUCUAUUGCCACGUUGUUGA-AAUCUAUUAGCCACAUUUAGUGUGGCUCAUAAAAUUCUAUUGCAUCAUUUGUUUCU--- ..((((.((---(.......))).)))).((.((.((-(.(.(((.((((((((...)))))))).))).).))).)).)).............--- ( -19.10, z-score = -2.47, R) >droYak2.chr3L 21568730 90 - 24197627 UUUGGCUGU---AUCUUUUCUAUUGCCACGUUGUUGA-AAUCUAUUAGCCACAUUUAGUGUGGCUCAUAAAAUUCUAUUGCAUCAUUUGUUUCU--- ..((((.((---(.......))).)))).((.((.((-(.(.(((.((((((((...)))))))).))).).))).)).)).............--- ( -19.10, z-score = -2.47, R) >droEre2.scaffold_4784 21011602 90 - 25762168 UUUGGCUGU---AUCUUUUCUAUUGCCACGUUGUUGA-AAUCUAUUAGCCACAUUUAGUGUGGCUCAUAAAAUUCUAUUGCAUCAUUUGUUUCU--- ..((((.((---(.......))).)))).((.((.((-(.(.(((.((((((((...)))))))).))).).))).)).)).............--- ( -19.10, z-score = -2.47, R) >droAna3.scaffold_13337 19687629 90 + 23293914 UUUGGCUGU---AUCUUUUCUAUUGCCACGUUGUUGA-AAUCUAUUAGCCACAUUUAGUGUGGCUCAUAAAAUUCUAUUGCAUCAUUUGUUCUU--- ..((((.((---(.......))).)))).((.((.((-(.(.(((.((((((((...)))))))).))).).))).)).)).............--- ( -19.10, z-score = -2.47, R) >dp4.chrXR_group5 502545 97 - 740492 UUUGGCUGUAGUAUCUUUUCUAUUGCCACGUUGUUGAGAAUCUAUUAGCCACAUUUAGUGUGGCUCAUAAAAUUCUAUUGCAUCAUUUGUUCUCUCU ..((((.((((........)))).)))).(.(((..(((((.(((.((((((((...)))))))).)))..)))))...))).)............. ( -26.10, z-score = -4.06, R) >droPer1.super_58 176761 97 + 410386 UUUGGCUGUAGUAUCUUUUCUAUUGCCACGUUGUUGAGAAUCUAUUAGCCACAUUUAGUGUGGCUCAUAAAAUUCUAUUGCAUCAUUUGUUCUCUCU ..((((.((((........)))).)))).(.(((..(((((.(((.((((((((...)))))))).)))..)))))...))).)............. ( -26.10, z-score = -4.06, R) >droWil1.scaffold_181024 561472 90 - 1751709 CCUGGCUGU---AUCUUUUCUAUUGCCACGUUGUUGA-AAUCUAUUAGCCACAUUUAGUGUGGCUCAUAAAAUUCUAUUGCAUCAUUUGUUUCU--- ..((((.((---(.......))).)))).((.((.((-(.(.(((.((((((((...)))))))).))).).))).)).)).............--- ( -19.00, z-score = -2.46, R) >droVir3.scaffold_13049 918648 92 + 25233164 GCUGUCAGU---AUCUUUUCUAUUGCCACAUUGUUAA-AAUCUAUUAGCCACAUUAAAUGUGGCUCGUAAAAUUCGAUUGCAUCAUUUGUUUCUCU- ((.(((.((---(..........)))...........-....(((.((((((((...)))))))).)))......))).))...............- ( -15.80, z-score = -1.89, R) >droMoj3.scaffold_6680 24404933 84 - 24764193 GCUGUCUGU---AUCUUUUC------CACAUUGUUAA-AAUCUAUUAGCCACAUUAAAUGUGGCUCGUAAAAUUCGAUUGCAUCAUUUGUUUCU--- ((.((((((---........------.))).......-....(((.((((((((...)))))))).)))......))).)).............--- ( -14.60, z-score = -2.18, R) >consensus UUUGGCUGU___AUCUUUUCUAUUGCCACGUUGUUGA_AAUCUAUUAGCCACAUUUAGUGUGGCUCAUAAAAUUCUAUUGCAUCAUUUGUUUCU___ ..((((..................))))..............(((.((((((((...)))))))).)))............................ (-13.95 = -13.92 + -0.02)
| Location | 9,294,876 – 9,294,966 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 91.10 |
| Shannon entropy | 0.18343 |
| G+C content | 0.33881 |
| Mean single sequence MFE | -19.80 |
| Consensus MFE | -15.59 |
| Energy contribution | -15.90 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.993186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
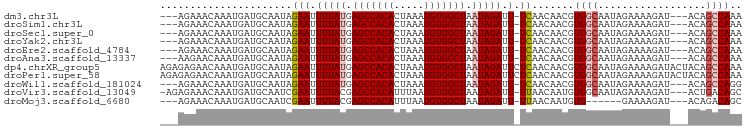
>dm3.chr3L 9294876 90 - 24543557 ---AGAAACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUU-UCAACAACGUGGCAAUAGAAAAGAU---ACAGCCAAA ---...................(((.(((((.(((((((.....))))))).))))).)-)).......((((..((.......)---)..)))).. ( -20.30, z-score = -3.65, R) >droSim1.chr3L 8684490 90 - 22553184 ---AGAAACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUU-UCAACAACGUGGCAAUAGAAAAGAU---ACAGCCAAA ---...................(((.(((((.(((((((.....))))))).))))).)-)).......((((..((.......)---)..)))).. ( -20.30, z-score = -3.65, R) >droSec1.super_0 1543806 90 - 21120651 ---AGAAACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUU-UCAACAACGUGGCAAUAGAAAAGAU---ACAGCCAAA ---...................(((.(((((.(((((((.....))))))).))))).)-)).......((((..((.......)---)..)))).. ( -20.30, z-score = -3.65, R) >droYak2.chr3L 21568730 90 + 24197627 ---AGAAACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUU-UCAACAACGUGGCAAUAGAAAAGAU---ACAGCCAAA ---...................(((.(((((.(((((((.....))))))).))))).)-)).......((((..((.......)---)..)))).. ( -20.30, z-score = -3.65, R) >droEre2.scaffold_4784 21011602 90 + 25762168 ---AGAAACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUU-UCAACAACGUGGCAAUAGAAAAGAU---ACAGCCAAA ---...................(((.(((((.(((((((.....))))))).))))).)-)).......((((..((.......)---)..)))).. ( -20.30, z-score = -3.65, R) >droAna3.scaffold_13337 19687629 90 - 23293914 ---AAGAACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUU-UCAACAACGUGGCAAUAGAAAAGAU---ACAGCCAAA ---...................(((.(((((.(((((((.....))))))).))))).)-)).......((((..((.......)---)..)))).. ( -20.30, z-score = -3.65, R) >dp4.chrXR_group5 502545 97 + 740492 AGAGAGAACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUUCUCAACAACGUGGCAAUAGAAAAGAUACUACAGCCAAA ...........(((.((....(((((((....(((((((.....)))))))...)))))))...)).)))(((..(((........)))..)))... ( -22.20, z-score = -3.47, R) >droPer1.super_58 176761 97 - 410386 AGAGAGAACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUUCUCAACAACGUGGCAAUAGAAAAGAUACUACAGCCAAA ...........(((.((....(((((((....(((((((.....)))))))...)))))))...)).)))(((..(((........)))..)))... ( -22.20, z-score = -3.47, R) >droWil1.scaffold_181024 561472 90 + 1751709 ---AGAAACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUU-UCAACAACGUGGCAAUAGAAAAGAU---ACAGCCAGG ---...................(((.(((((.(((((((.....))))))).))))).)-)).....(.((((..((.......)---)..)))).) ( -21.30, z-score = -3.75, R) >droVir3.scaffold_13049 918648 92 - 25233164 -AGAGAAACAAAUGAUGCAAUCGAAUUUUACGAGCCACAUUUAAUGUGGCUAAUAGAUU-UUAACAAUGUGGCAAUAGAAAAGAU---ACUGACAGC -...(..(((...(((...)))(((.((((..((((((((...))))))))..)))).)-)).....)))..)............---......... ( -14.80, z-score = -1.01, R) >droMoj3.scaffold_6680 24404933 84 + 24764193 ---AGAAACAAAUGAUGCAAUCGAAUUUUACGAGCCACAUUUAAUGUGGCUAAUAGAUU-UUAACAAUGUG------GAAAAGAU---ACAGACAGC ---.............((..(((((.((((..((((((((...))))))))..)))).)-)).....((((------.......)---)))))..)) ( -15.50, z-score = -2.18, R) >consensus ___AGAAACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUU_UCAACAACGUGGCAAUAGAAAAGAU___ACAGCCAAA ......................((..(((((.(((((((.....))))))).)))))...)).......((((..................)))).. (-15.59 = -15.90 + 0.31)
| Location | 9,294,880 – 9,294,973 |
|---|---|
| Length | 93 |
| Sequences | 11 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 90.50 |
| Shannon entropy | 0.19574 |
| G+C content | 0.34598 |
| Mean single sequence MFE | -18.80 |
| Consensus MFE | -13.53 |
| Energy contribution | -13.84 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.866043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
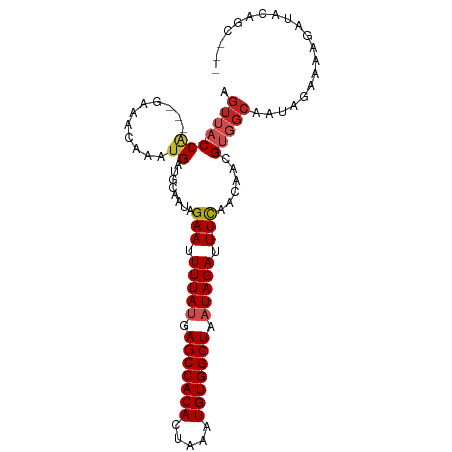
>dm3.chr3L 9294880 93 - 24543557 AGUUA-CCA---GAAACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUU-UCAACAACGUGGCAAUAGAAAAGAUACAGC--- .((((-(((---........))........(((.(((((.(((((((.....))))))).))))).)-))......))))).................--- ( -19.90, z-score = -2.96, R) >droSim1.chr3L 8684494 93 - 22553184 AGUUA-CCA---GAAACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUU-UCAACAACGUGGCAAUAGAAAAGAUACAGC--- .((((-(((---........))........(((.(((((.(((((((.....))))))).))))).)-))......))))).................--- ( -19.90, z-score = -2.96, R) >droSec1.super_0 1543810 93 - 21120651 AGUUA-CCA---GAAACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUU-UCAACAACGUGGCAAUAGAAAAGAUACAGC--- .((((-(((---........))........(((.(((((.(((((((.....))))))).))))).)-))......))))).................--- ( -19.90, z-score = -2.96, R) >droYak2.chr3L 21568734 93 + 24197627 AGUUA-CCA---GAAACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUU-UCAACAACGUGGCAAUAGAAAAGAUACAGC--- .((((-(((---........))........(((.(((((.(((((((.....))))))).))))).)-))......))))).................--- ( -19.90, z-score = -2.96, R) >droEre2.scaffold_4784 21011606 93 + 25762168 AGUUA-CCA---GAAACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUU-UCAACAACGUGGCAAUAGAAAAGAUACAGC--- .((((-(((---........))........(((.(((((.(((((((.....))))))).))))).)-))......))))).................--- ( -19.90, z-score = -2.96, R) >droAna3.scaffold_13337 19687633 93 - 23293914 AGUUA-CCA---AGAACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUU-UCAACAACGUGGCAAUAGAAAAGAUACAGC--- .((((-(((---........))........(((.(((((.(((((((.....))))))).))))).)-))......))))).................--- ( -19.10, z-score = -2.64, R) >dp4.chrXR_group5 502549 100 + 740492 AGUUA-CCAGAGAGAACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUUCUCAACAACGUGGCAAUAGAAAAGAUACUACAGC .((((-(....((((((....)............(((((.(((((((.....))))))).))))))))))......))))).................... ( -18.50, z-score = -1.58, R) >droPer1.super_58 176765 100 - 410386 AGUUA-CCAGAGAGAACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUUCUCAACAACGUGGCAAUAGAAAAGAUACUACAGC .((((-(....((((((....)............(((((.(((((((.....))))))).))))))))))......))))).................... ( -18.50, z-score = -1.58, R) >droWil1.scaffold_181024 561476 94 + 1751709 AGUUACCCA---GAAACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUU-UCAACAACGUGGCAAUAGAAAAGAUACAGC--- .(((((.((---........))........(((.(((((.(((((((.....))))))).))))).)-))......))))).................--- ( -19.90, z-score = -2.82, R) >droVir3.scaffold_13049 918652 95 - 25233164 AGUA--CCGAGAGAAACAAAUGAUGCAAUCGAAUUUUACGAGCCACAUUUAAUGUGGCUAAUAGAUU-UUAACAAUGUGGCAAUAGAAAAGAUACUGA--- ((((--.(....)..........(((....(((.((((..((((((((...))))))))..)))).)-))..(.....))))..........))))..--- ( -16.00, z-score = -1.22, R) >droMoj3.scaffold_6680 24404937 87 + 24764193 AGUA--CC--GAGAAACAAAUGAUGCAAUCGAAUUUUACGAGCCACAUUUAAUGUGGCUAAUAGAUU-UUAACAAUGUG------GAAAAGAUACAGA--- .(((--.(--.....(((...(((...)))(((.((((..((((((((...))))))))..)))).)-)).....))).------.....).)))...--- ( -15.30, z-score = -1.69, R) >consensus AGUUA_CCA___GAAACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUU_UCAACAACGUGGCAAUAGAAAAGAUACAGC___ .......................(((....((..(((((.(((((((.....))))))).)))))...))..(.....))))................... (-13.53 = -13.84 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:13:29 2011