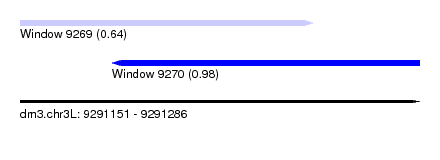
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,291,151 – 9,291,286 |
| Length | 135 |
| Max. P | 0.980628 |

| Location | 9,291,151 – 9,291,250 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 72.59 |
| Shannon entropy | 0.47492 |
| G+C content | 0.41251 |
| Mean single sequence MFE | -25.91 |
| Consensus MFE | -11.94 |
| Energy contribution | -12.48 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.641786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
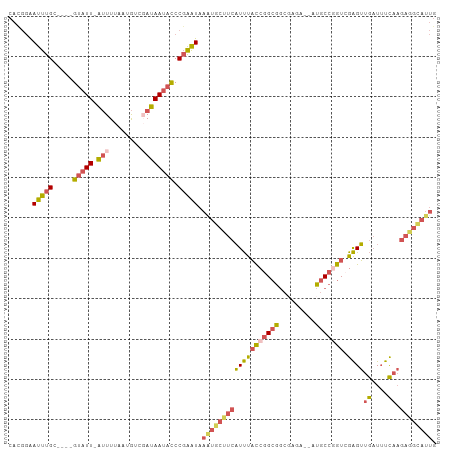
>dm3.chr3L 9291151 99 + 24543557 CAUGGAAUUUGC----GUAUU-AUUUUAAUGUCGAUAAUACCCGAAUAAAUGCCUCAUUUACCGGCGGCGAGA--AUGCCAGUGGAGUUGAUUUCAAGAGGCAUUG ......(((((.----(((((-(((........)))))))).))))).((((((((.(((((.((((......--.)))).))))).(((....))))))))))). ( -29.80, z-score = -2.75, R) >droSim1.chr3L 8680724 99 + 22553184 CACGGAAUUUGC----GUAUU-AUUUUAAUGUCGAUAAUACCCGAAUAAAUGCUUCAUUUACCGGCGGCGAGA--AUGCCGGUCGAGUUGAUUUCAAGAGGUAUUG ......(((((.----(((((-(((........)))))))).))))).((((((((....(((((((......--.))))))).(((.....)))..)))))))). ( -28.10, z-score = -2.05, R) >droSec1.super_0 1540055 99 + 21120651 CACGGAAUUUGC----GUAUU-AUUUUAAUGUCGAUAAUACCCGAAUAAGUGCUUCAUUUACCGGCGGCGGGA--UUGCCGGUCGAGUUGAUUUCAAGAGGCAUUG ......(((((.----(((((-(((........)))))))).))))).((((((((....((((((((.....--)))))))).(((.....)))..)))))))). ( -30.10, z-score = -2.04, R) >droEre2.scaffold_4784 21007983 99 - 25762168 CACGGAAUUUGC----GUAUU-AUUUUAAUGUCCAUAAUUCCCGAAUAAAUGCUUCAUUUACCGGCGGCCAGA--AUGCCGGUCGAGUUGAUUUCAAGAGGCAUUG ...((((((((.----.((((-(...)))))..)).))))))......((((((((....(((((((......--.))))))).(((.....)))..)))))))). ( -28.20, z-score = -2.17, R) >droAna3.scaffold_13337 19683902 93 + 23293914 UUCUCGAUUCGCCAUUGUAUUUGCUUUAUAAAAUAUAAAAUCCCGGUAAUU--UUCCUCCCUCAGCACACAGAUUAUACCAAUGGGGGAACUAUU----------- ..............((((((((........))))))))......((.....--..))(((((((..................)))))))......----------- ( -13.37, z-score = 0.41, R) >consensus CACGGAAUUUGC____GUAUU_AUUUUAAUGUCGAUAAUACCCGAAUAAAUGCUUCAUUUACCGGCGGCGAGA__AUGCCGGUCGAGUUGAUUUCAAGAGGCAUUG ................................................(((((((((((((((((((.........))))))).))))((....)).)))))))). (-11.94 = -12.48 + 0.54)

| Location | 9,291,182 – 9,291,286 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 69.63 |
| Shannon entropy | 0.58895 |
| G+C content | 0.41348 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -13.54 |
| Energy contribution | -14.50 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.980628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
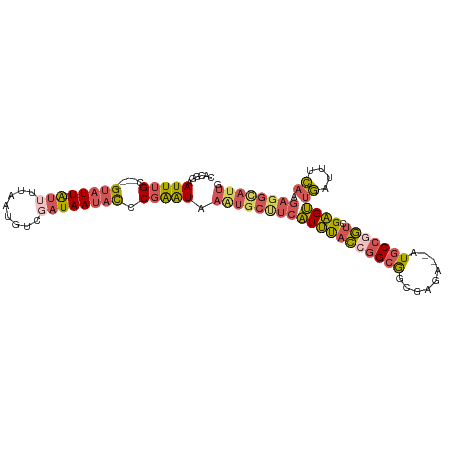
>dm3.chr3L 9291182 104 - 24543557 UAAUUGGCGAGAUUUCUGAUUUAUUCAGUAAUUUUGCAAUGCCUCUUGAAAUCAACUCCACUGGCAUUCUCGCCGCCGGUAAAUGAGGCAUUUAUUCGGGUAUU ......((((((((.((((.....)))).))))))))(((((((((((....)))....((((((.........))))))....))))))))............ ( -32.10, z-score = -3.14, R) >droSim1.chr3L 8680755 104 - 22553184 UAAGUGGCGAGAUUUCUGAUUCAUUCAGUAAUUUUGCAAUACCUCUUGAAAUCAACUCGACCGGCAUUCUCGCCGCCGGUAAAUGAAGCAUUUAUUCGGGUAUU ......((((((((.((((.....)))).))))))))(((((((....(((((..(...((((((.........))))))....)..).))))....))))))) ( -29.90, z-score = -2.57, R) >droSec1.super_0 1540086 104 - 21120651 UAAGUGGCGAGAUUUCUGAUUCAUUCAGUAAUUUUGCAAUGCCUCUUGAAAUCAACUCGACCGGCAAUCCCGCCGCCGGUAAAUGAAGCACUUAUUCGGGUAUU ((((((((((((((.((((.....)))).))))))))..............(((.....((((((.........))))))...)))..)))))).......... ( -29.50, z-score = -1.92, R) >droYak2.chr3L 21564965 81 + 24197627 UAAGUGGCGAGAUUACGGAUUCAUUCAGUGAUUUUGCAAUGCCUCUUGAAAUCAACUCAA-UGAAGCAUUUAUUGCCG--AAUU-------------------- ......((((((((((.((.....)).))))))))))(((((.((((((.......))))-.)).)))))........--....-------------------- ( -19.80, z-score = -1.61, R) >droEre2.scaffold_4784 21008014 104 + 25762168 UAAGUGGCGAGAUUUCUGAUUCAUUCGGUCAUUUUGCAAUGCCUCUUGAAAUCAACUCGACCGGCAUUCUGGCCGCCGGUAAAUGAAGCAUUUAUUCGGGAAUU ......(((((((..((((.....))))..)))))))......(((((((.........((((((.........))))))(((((...))))).)))))))... ( -27.20, z-score = -0.42, R) >droAna3.scaffold_13337 19683938 93 - 23293914 CAAGUGCCUCGAAUUCUGAUUUAUUCUGUGAUUUCG-AAUAGUUCCC-CCAUUGGUAUAAUCUGUGUGCUGAGGGAGGAAAAUUACCGGGAUUUU--------- .((((.(((((((.((.((.....))...)).))))-)....(((((-((.(..(((((.....)))))..)))).)))).......)).)))).--------- ( -25.90, z-score = -1.78, R) >consensus UAAGUGGCGAGAUUUCUGAUUCAUUCAGUAAUUUUGCAAUGCCUCUUGAAAUCAACUCGACCGGCAUUCUCGCCGCCGGUAAAUGAAGCAUUUAUUCGGGUAUU ......((((((((.((((.....)))).))))))))......................((((((.........))))))........................ (-13.54 = -14.50 + 0.96)
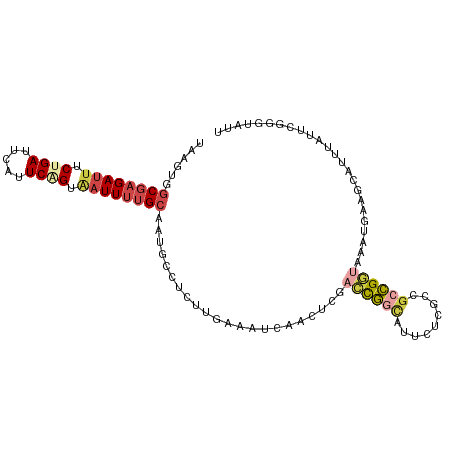
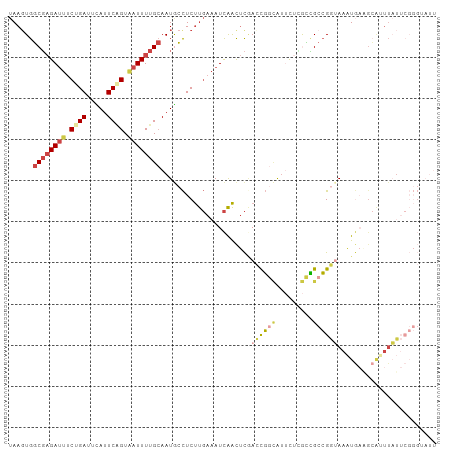
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:13:27 2011