| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,286,631 – 9,286,728 |
| Length | 97 |
| Max. P | 0.938858 |

| Location | 9,286,631 – 9,286,728 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 76.21 |
| Shannon entropy | 0.46395 |
| G+C content | 0.34001 |
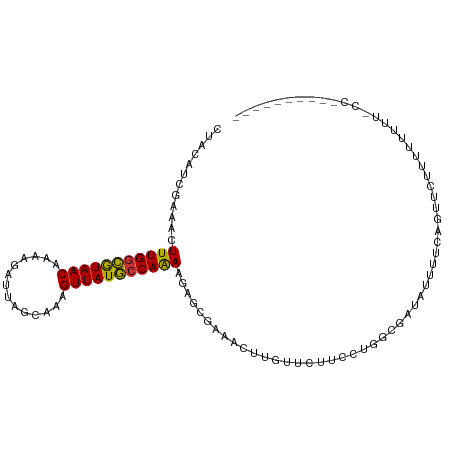
| Mean single sequence MFE | -18.59 |
| Consensus MFE | -12.29 |
| Energy contribution | -11.98 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.938858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
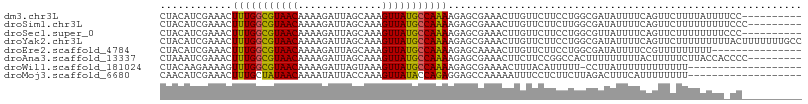
>dm3.chr3L 9286631 97 + 24543557 CUACAUCGAAACUUUGGCGUAACAAAAGAUUAGCAAAGUUAUGCCAAAAGAGCGAAACUUGUUCUUCCUGGCGAUAUUUUCAGUUCUUUUAUUUUCC---------- .......((((....((((((((..............))))))))((((((((((((.(((((......)))))...)))).))))))))..)))).---------- ( -20.94, z-score = -1.69, R) >droSim1.chr3L 8676186 98 + 22553184 CUACAUCGAAACUUUGGCGUAACAAAAGAUUAGCAAAGUUAUGCCAAAAGAGCGAAACUUGUUCUUCUUGGCGAUAUUUUCAGUUCUUUUUUUUUCCC--------- .......((((....((((((((..............))))))))((((((((((((.(((((......)))))...)))).))))))))..))))..--------- ( -20.34, z-score = -1.31, R) >droSec1.super_0 1535600 97 + 21120651 CUACAUCGAAACUUUGGCGUAACAAAAGAUUAGCAAAGUUAUGCCAAAAGAGCGAAACUUGUUCUUCCUGGCGUUAUUUUCAGUUCUUUUUUUUCCC---------- .(((..(((....)))..)))..((((((...(.(((((.((((((.((((((((...))))))))..)))))).))))))...)))))).......---------- ( -21.00, z-score = -1.63, R) >droYak2.chr3L 21559333 107 - 24197627 CUACAUCGAAACUUUGGCGUAACAAAAGAUUAGCAAAGUUAUGCCAAAAGAGCGAAACUUGUUCUUCCUGGCGAUAUUUUCAGUUCUUUUUUUUUACUUUUUUUGCC ....((((.....((((((((((..............))))))))))((((((((...)))))))).....))))................................ ( -20.04, z-score = -0.60, R) >droEre2.scaffold_4784 21003449 92 - 25762168 CUACAUCGAAACUUUGGCGUAACAAAAGAUUAGCAAAGUUAUGCCAAAAGAGCAAAACUUGUUCUUCCUGGCGAUAUUUUCCGUUUUUUUUU--------------- ....((((.....((((((((((..............))))))))))(((((((.....))))))).....)))).................--------------- ( -20.34, z-score = -1.90, R) >droAna3.scaffold_13337 19678974 98 + 23293914 CUAAAUCGAAACUUUGGCGUAACAAAAGAUUAGCAAAGUUAUGCCAAAAGAGCGAAACUUCUUCCGGCCACUUUUUUUUUACUUUUUCUUACCACCCC--------- .....(((....(((((((((((..............)))))))))))....)))...........................................--------- ( -15.94, z-score = -0.89, R) >droWil1.scaffold_181024 539143 87 - 1751709 CUACAAGAAAAGUUUGGCGUAACAAAAGAUUAGUAAAGUUAUGCCAAAAGAGCGAAAACUUUACAUUUUU-CCUUAUUUUUUUUUUUU------------------- ....(((((((((((((((((((..............))))))))))).(((.(((((........))))-)))).))))))))....------------------- ( -17.84, z-score = -2.29, R) >droMoj3.scaffold_6680 24394570 88 - 24764193 CAACAUCGAAACUUUGCUAUAACAAAAUAUUACCAAAGUUAUACCAGAGGAGCCAAAAAUUUCCUCUUCUUAGACUUUCAUUUUUUUU------------------- .......(((((((((..(((......)))...))))))).....(((((((........)))))))..........)).........------------------- ( -12.30, z-score = -2.31, R) >consensus CUACAUCGAAACUUUGGCGUAACAAAAGAUUAGCAAAGUUAUGCCAAAAGAGCGAAACUUGUUCUUCCUGGCGAUAUUUUCAGUUCUUUUUUUU_CC__________ ............(((((((((((..............)))))))))))........................................................... (-12.29 = -11.98 + -0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:13:23 2011