| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,259,285 – 9,259,357 |
| Length | 72 |
| Max. P | 0.624816 |

| Location | 9,259,285 – 9,259,357 |
|---|---|
| Length | 72 |
| Sequences | 7 |
| Columns | 89 |
| Reading direction | forward |
| Mean pairwise identity | 68.18 |
| Shannon entropy | 0.57707 |
| G+C content | 0.42478 |
| Mean single sequence MFE | -22.51 |
| Consensus MFE | -5.64 |
| Energy contribution | -5.67 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.62 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.624816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9259285 72 + 24543557 --------GCUAAUGCAUCU----AUAUAUCUGGU-UCUGGAUCUG---GUCAUAUGACCGGAUGGCC-CAGAAUGUGUGUAAAUUUGC --------((....))...(----((((((...((-((((((((((---(((....))))))))...)-)))))))))))))....... ( -24.80, z-score = -3.28, R) >droSim1.chr3L 8656705 72 + 22553184 --------GCUAAUGCAUCU----AUAUAUCUGGA-UCUGGAUCUG---GUCAUAUGACCGGAUGGCC-CAGAAUGUGUGUAAAUUUGC --------((....))...(----((((((((((.-.(...(((((---(((....)))))))))..)-))).)))))))......... ( -23.70, z-score = -2.77, R) >droSec1.super_0 1516062 72 + 21120651 --------GCUAAUGCAUCU----AUAUAUCUGGA-UCUGGAUCUG---GUCAUAUGACCGGAUAGCC-CAGAAUGUGUGUAAAUUUGC --------((....))...(----((((((((((.-.....(((((---(((....))))))))...)-))).)))))))......... ( -22.30, z-score = -2.52, R) >droEre2.scaffold_4784 20983987 72 - 25762168 --------GCUAAUGCAUCU----AUAUAUCUGGA-UCUGGAUCUG---GUCAUAUGACCGGAUGGCC-CAGAAUGUGUGUAAAUUUGC --------((....))...(----((((((((((.-.(...(((((---(((....)))))))))..)-))).)))))))......... ( -23.70, z-score = -2.77, R) >droYak2.chr3L 21538874 76 - 24197627 --------GCUAAUGCAUAUCCAUAUAUAUCUGGA-UCUGGAUCUG---GUCAUAAGACCGGAUGGGC-CAGAAUGUGUGUAAAUUUGC --------((....)).....(((((((.(((((.-(((..(((((---(((....))))))))))))-)))))))))))......... ( -26.70, z-score = -3.20, R) >droAna3.scaffold_13337 19659880 89 + 23293914 UCCAAUGCAUAUGUAUGUGUGGCCAUAUGGCCAUUGUCUGUGUCCGUGUGUGAGAGAGUCUGACGGGAGCGGGGGGUGUGUAAAUUUAC .....(((((((.(....((((((....))))))........(((((.(((.((.....)).)))...)))))).)))))))....... ( -25.70, z-score = -0.36, R) >droGri2.scaffold_15074 4402877 65 - 7742996 --------GUUUCAGGGCAA----AUAUAUCUCGU-CCUGCAGCAC---AUUAUAUAACCGGAUA-----AAAACGAUAAAAAAUU--- --------(((.((((((..----.........))-)))).)))..---..........((....-----....))..........--- ( -10.70, z-score = -1.92, R) >consensus ________GCUAAUGCAUCU____AUAUAUCUGGA_UCUGGAUCUG___GUCAUAUGACCGGAUGGCC_CAGAAUGUGUGUAAAUUUGC ............((((((.........(((((((......(((......)))......)))))))..........))))))........ ( -5.64 = -5.67 + 0.03)

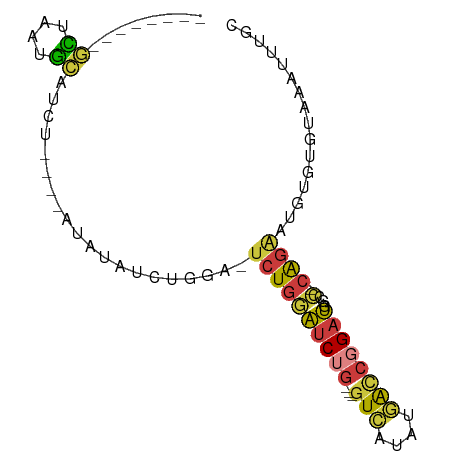
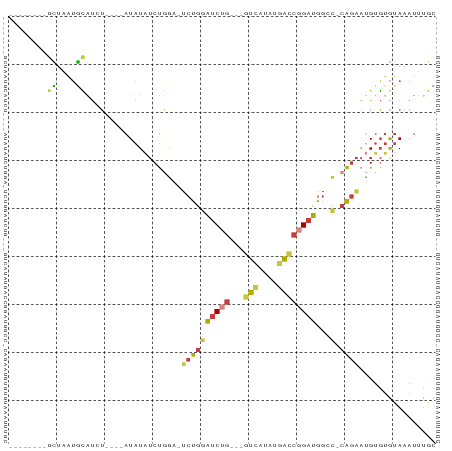
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:13:22 2011