| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,646,100 – 4,646,163 |
| Length | 63 |
| Max. P | 0.656380 |

| Location | 4,646,100 – 4,646,163 |
|---|---|
| Length | 63 |
| Sequences | 6 |
| Columns | 81 |
| Reading direction | forward |
| Mean pairwise identity | 65.31 |
| Shannon entropy | 0.57165 |
| G+C content | 0.57349 |
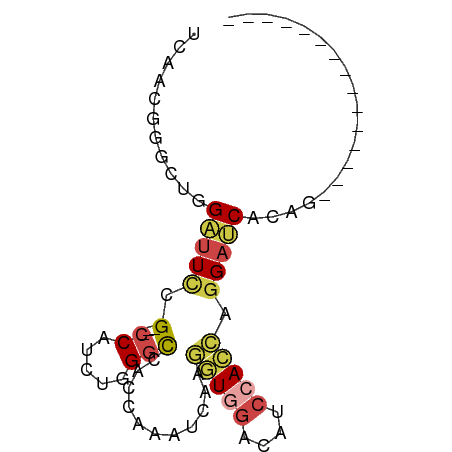
| Mean single sequence MFE | -20.77 |
| Consensus MFE | -8.24 |
| Energy contribution | -8.97 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.656380 |
| Prediction | RNA |
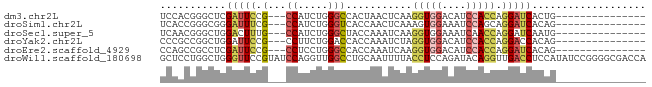
Download alignment: ClustalW | MAF
>dm3.chr2L 4646100 63 + 23011544 UCCACGGGCUCGAUUCCG---CCAUCUGGGCCACUAACUCAAGGUGGACAUCCACCAGGAUCACUG--------------- (((...((((((((....---..))).)))))..........(((((....))))).)))......--------------- ( -21.20, z-score = -2.20, R) >droSim1.chr2L 4518553 63 + 22036055 UCACCGGGCGGGAUUUCG---CCAUCUGGGUCACCAACUCAAAGUGGAAAUCCAGCAGGAUCACAG--------------- ...((..((.(((((...---((((.(((((.....)))))..)))).))))).)).)).......--------------- ( -18.40, z-score = -1.46, R) >droSec1.super_5 2730295 63 + 5866729 UCAACGGGCUGGACUUUG---CCAUCUGGGCUACCAAAUCAAGGUGGAAAUCAACCAGGAUCAAUG--------------- ......(((........)---)).(((((.(((((.......))))).......))))).......--------------- ( -15.50, z-score = -0.70, R) >droYak2.chr2L 4659006 63 + 22324452 CCCGCCGGCUGGAUUCCG---CCUUCUGGACCACCAAAUCUAGGUGGACAUCCACCAGGACCACAG--------------- ....((((.(((((((((---(((..(((....))).....))))))).))))))).)).......--------------- ( -22.40, z-score = -2.39, R) >droEre2.scaffold_4929 4724948 63 + 26641161 CCAGCCGCCUCGAUUCCG---CCUCCUGGGCCACCAAAUCAAGGUGGACAUCCACCAGGAUCACAG--------------- ...(((((((.((((..(---((.....))).....)))).)))))).)((((....)))).....--------------- ( -18.50, z-score = -1.58, R) >droWil1.scaffold_180698 4447202 81 - 11422946 GCUCCUGGCUGGGUUCCGUAUCCAGGUUGGCCUGCAAUUUUACCUCCAGAUACAGGUUGACCUCCAUAUCCGGGGCGACCA ((.(((((.(((((.....)))))(((..(((((..((((.......)))).)))))..))).......)))))))..... ( -28.60, z-score = -1.08, R) >consensus UCAACGGGCUGGAUUCCG___CCAUCUGGGCCACCAAAUCAAGGUGGACAUCCACCAGGAUCACAG_______________ ...........(((((..........(((....)))......(((((....))))).)))))................... ( -8.24 = -8.97 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:17:08 2011