| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,213,090 – 9,213,185 |
| Length | 95 |
| Max. P | 0.934866 |

| Location | 9,213,090 – 9,213,185 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 89.83 |
| Shannon entropy | 0.21432 |
| G+C content | 0.50317 |
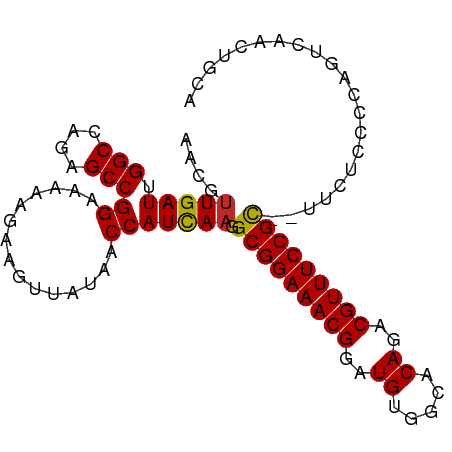
| Mean single sequence MFE | -33.05 |
| Consensus MFE | -22.35 |
| Energy contribution | -22.15 |
| Covariance contribution | -0.20 |
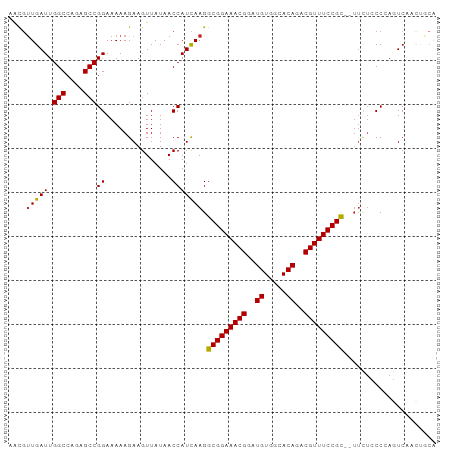
| Combinations/Pair | 1.10 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
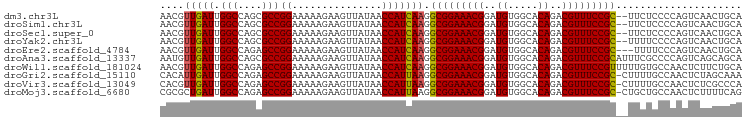
>dm3.chr3L 9213090 95 + 24543557 AACGUUGAUUGGCCAGCGCCGGAAAAAGAAGUUAUAACCAUCAAGGCGGAAACGGAUGUGGCACAGACGUUUCCGC--UUCUCCCCAGUCAACUGCA ...(((((((((((......)).....................(((((((((((..((.....))..)))))))))--))....))))))))).... ( -33.30, z-score = -2.62, R) >droSim1.chr3L 8610161 95 + 22553184 AACGUUGAUUGGCCAGCGCCGGAAAAAGAAGUUAUAACCAUCAAGGCGGAAACGGAUGUGGCACAGACGUUUCCGC--UUCUCCCCAGUCAACUGCA ...(((((((((((......)).....................(((((((((((..((.....))..)))))))))--))....))))))))).... ( -33.30, z-score = -2.62, R) >droSec1.super_0 1469378 95 + 21120651 AACGUUGAUUGGCCAGCGCCGGAAAAAGAAGUUAUAACCAUCAAGGCGGAAACGGAUGUGGCACAGACGUUUCCGC--UUCUCCCCAGUCAACUGCA ...(((((((((((......)).....................(((((((((((..((.....))..)))))))))--))....))))))))).... ( -33.30, z-score = -2.62, R) >droYak2.chr3L 21491399 95 - 24197627 AACGUUGAUUGGCCAGCGCCGGAAAAAGAAGUUAUAACCAUCAAGGCGGAAACGGAUGUGGCACAGACGUUUCCGC--UUUUCCCCAGUCAACUGCA ...(((((((((((......))....................((((((((((((..((.....))..)))))))))--)))...))))))))).... ( -34.20, z-score = -3.02, R) >droEre2.scaffold_4784 20931931 94 - 25762168 AACGUUGAUUGGCCAGAGCCGGAAAAAGAAGUUAUAACCAUCAAGGCGGAAACGGAUGUGGCACAGACGUUUCCGC---UUUUCCCAGUCAACUGCA ...(((((((((((......))....................((((((((((((..((.....))..)))))))))---)))..))))))))).... ( -34.20, z-score = -3.40, R) >droAna3.scaffold_13337 19613364 97 + 23293914 AAUGUUGAUUGGCCAGCGCCGGAAAAAGAAGUUAUAACCAUCAAGGCGGAAACGGAUGUGGCACAGACGUUUCCGCAUUUCGCCCCAGUCAGCAGCA ..((((((((((...(((..((...............))......(((((((((..((.....))..)))))))))....))).))))))))))... ( -35.26, z-score = -3.11, R) >droWil1.scaffold_181024 1298824 97 + 1751709 AACGUUGAUUGGCCAGAGCCGGAAAAAGAAGUUAUAACCAUCAAGGCGGAAACGGAUGUGGCACAGACGUUUCCGUUUUUGUGCCAACUCUUCUGCA ........(((((....)))))....(((((.......(((.((((((((((((..((.....))..)))))))))))).)))......)))))... ( -30.32, z-score = -1.77, R) >droGri2.scaffold_15110 2017691 96 + 24565398 CACAUUGAUUGGCCAGAGCCGGAAAAAGAAGUUAUAACCAUUAAGGCGGAAACGGAUGUGGCACAGACGUUUCCGC-CUUUUGCCAACUCUAGCAAA ...........((.((((..((....................((((((((((((..((.....))..)))))))))-)))...))..)))).))... ( -30.80, z-score = -2.96, R) >droVir3.scaffold_13049 811130 96 + 25233164 CACGUUGAUUGGCCAGAGCCGGAAAAAGAAGUUAUAACCAUUAAGGCGGAAACGGAUGUGGCACAGACGUUUCCGC-CUUUUGCCAACUCUCGCCCA ..........(((.((((..((....................((((((((((((..((.....))..)))))))))-)))...))..)))).))).. ( -33.20, z-score = -2.75, R) >droMoj3.scaffold_6680 24318716 96 - 24764193 CGCGCUGAUUGGCCAGAGCCGGAAAAAGAAGUUAUAACCAUUAAGGCGGAAACGGAUGUGGCACAGACGUUUCCGC-CUGCUGCCAACUCUUUUCAG ....(((((((((....)))))..(((((.(((.....((...(((((((((((..((.....))..)))))))))-))..))..)))))))))))) ( -32.60, z-score = -2.16, R) >consensus AACGUUGAUUGGCCAGAGCCGGAAAAAGAAGUUAUAACCAUCAAGGCGGAAACGGAUGUGGCACAGACGUUUCCGC__UUCUCCCCAGUCAACUGCA ....(((((.(((....)))((...............))))))).(((((((((..((.....))..)))))))))..................... (-22.35 = -22.15 + -0.20)
| Location | 9,213,090 – 9,213,185 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 89.83 |
| Shannon entropy | 0.21432 |
| G+C content | 0.50317 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -20.95 |
| Energy contribution | -21.05 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.862686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9213090 95 - 24543557 UGCAGUUGACUGGGGAGAA--GCGGAAACGUCUGUGCCACAUCCGUUUCCGCCUUGAUGGUUAUAACUUCUUUUUCCGGCGCUGGCCAAUCAACGUU ....(((((...(((((((--(.(((((((..((.....))..)))))))(((.....)))........))))))))(((....)))..)))))... ( -32.50, z-score = -1.85, R) >droSim1.chr3L 8610161 95 - 22553184 UGCAGUUGACUGGGGAGAA--GCGGAAACGUCUGUGCCACAUCCGUUUCCGCCUUGAUGGUUAUAACUUCUUUUUCCGGCGCUGGCCAAUCAACGUU ....(((((...(((((((--(.(((((((..((.....))..)))))))(((.....)))........))))))))(((....)))..)))))... ( -32.50, z-score = -1.85, R) >droSec1.super_0 1469378 95 - 21120651 UGCAGUUGACUGGGGAGAA--GCGGAAACGUCUGUGCCACAUCCGUUUCCGCCUUGAUGGUUAUAACUUCUUUUUCCGGCGCUGGCCAAUCAACGUU ....(((((...(((((((--(.(((((((..((.....))..)))))))(((.....)))........))))))))(((....)))..)))))... ( -32.50, z-score = -1.85, R) >droYak2.chr3L 21491399 95 + 24197627 UGCAGUUGACUGGGGAAAA--GCGGAAACGUCUGUGCCACAUCCGUUUCCGCCUUGAUGGUUAUAACUUCUUUUUCCGGCGCUGGCCAAUCAACGUU ....(((((...(((((((--(.(((((((..((.....))..)))))))(((.....)))........))))))))(((....)))..)))))... ( -32.40, z-score = -2.14, R) >droEre2.scaffold_4784 20931931 94 + 25762168 UGCAGUUGACUGGGAAAA---GCGGAAACGUCUGUGCCACAUCCGUUUCCGCCUUGAUGGUUAUAACUUCUUUUUCCGGCUCUGGCCAAUCAACGUU ....(((((..(((((((---(.(((((((..((.....))..)))))))(((.....)))........))))))))(((....)))..)))))... ( -32.60, z-score = -3.01, R) >droAna3.scaffold_13337 19613364 97 - 23293914 UGCUGCUGACUGGGGCGAAAUGCGGAAACGUCUGUGCCACAUCCGUUUCCGCCUUGAUGGUUAUAACUUCUUUUUCCGGCGCUGGCCAAUCAACAUU .(((.(.....).))).....(((((((((..((.....))..))))))))).(((((((...............))(((....))).))))).... ( -27.86, z-score = -0.61, R) >droWil1.scaffold_181024 1298824 97 - 1751709 UGCAGAAGAGUUGGCACAAAAACGGAAACGUCUGUGCCACAUCCGUUUCCGCCUUGAUGGUUAUAACUUCUUUUUCCGGCUCUGGCCAAUCAACGUU ...(((((.((.((((((...(((....))).))))))))..(((((........)))))......)))))......(((....))).......... ( -28.40, z-score = -1.94, R) >droGri2.scaffold_15110 2017691 96 - 24565398 UUUGCUAGAGUUGGCAAAAG-GCGGAAACGUCUGUGCCACAUCCGUUUCCGCCUUAAUGGUUAUAACUUCUUUUUCCGGCUCUGGCCAAUCAAUGUG ...(((((((((((...(((-(((((((((..((.....))..))))))))))))...(((....))).......)))))))))))........... ( -38.80, z-score = -5.66, R) >droVir3.scaffold_13049 811130 96 - 25233164 UGGGCGAGAGUUGGCAAAAG-GCGGAAACGUCUGUGCCACAUCCGUUUCCGCCUUAAUGGUUAUAACUUCUUUUUCCGGCUCUGGCCAAUCAACGUG ..(((.((((((((...(((-(((((((((..((.....))..))))))))))))...(((....))).......)))))))).))).......... ( -40.90, z-score = -4.77, R) >droMoj3.scaffold_6680 24318716 96 + 24764193 CUGAAAAGAGUUGGCAGCAG-GCGGAAACGUCUGUGCCACAUCCGUUUCCGCCUUAAUGGUUAUAACUUCUUUUUCCGGCUCUGGCCAAUCAGCGCG ((((((((((.(((((.(((-(((....))))))))))).....(((...(((.....)))...)))))))))....(((....)))..)))).... ( -34.30, z-score = -2.79, R) >consensus UGCAGUUGACUGGGGAGAA__GCGGAAACGUCUGUGCCACAUCCGUUUCCGCCUUGAUGGUUAUAACUUCUUUUUCCGGCGCUGGCCAAUCAACGUU ............((.......(((((((((..((.....))..)))))))))......(((....))).......))(((....))).......... (-20.95 = -21.05 + 0.10)

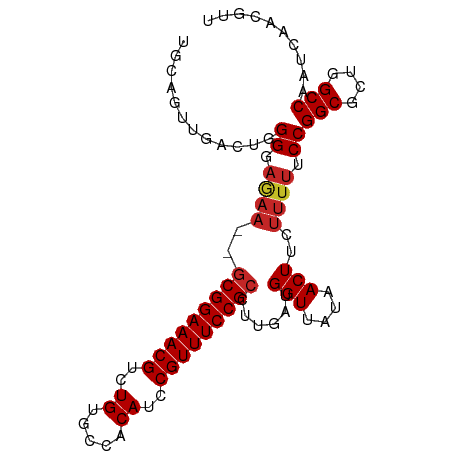
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:13:18 2011