| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,212,205 – 9,212,312 |
| Length | 107 |
| Max. P | 0.973284 |

| Location | 9,212,205 – 9,212,312 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.14 |
| Shannon entropy | 0.40074 |
| G+C content | 0.51978 |
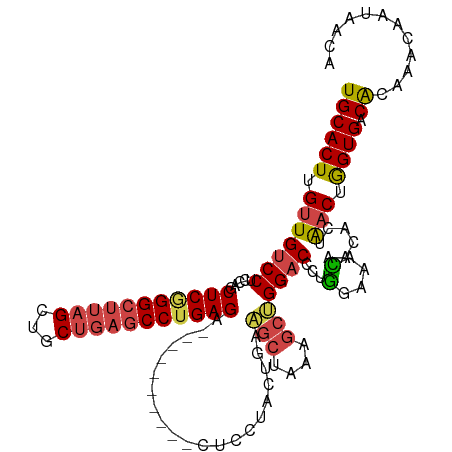
| Mean single sequence MFE | -37.67 |
| Consensus MFE | -22.32 |
| Energy contribution | -22.88 |
| Covariance contribution | 0.56 |
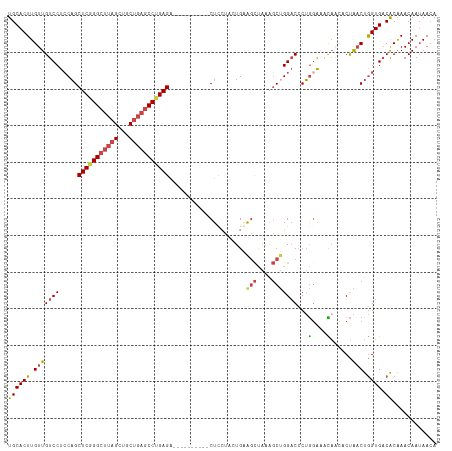
| Combinations/Pair | 1.25 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973284 |
| Prediction | RNA |
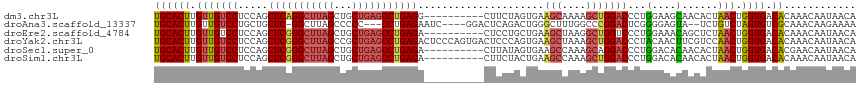
Download alignment: ClustalW | MAF
>dm3.chr3L 9212205 107 - 24543557 UGCACUUGUUGUCCUCCAGCUCAGGCUUAGCUGCUGAGCCUGAGG----------CUUCUAGUGAAGCAAAAGCUGGACCCUGGAAGCAACACUAACUGGUGACACAAACAAUAACA .....((((.(((..(((((((((((((((...)))))))))))(----------(((((((.(.(((....)))...).))))))))........)))).)))))))......... ( -43.10, z-score = -3.74, R) >droAna3.scaffold_13337 19613079 107 - 23293914 UGCACUUGUUGUCCUGCUGCUC-GGCUUAGCCCCC---CCUGAGAAUC----GGACUCAGACUGGGCUUUGGCCCGGACUCGGGGAGUA--UCUGUCUAGUGUCGCAAACAAGAAAA ....((((((((..(((((..(-((.....((((.---.(((((....----...))))).((((((....))))))....))))....--.)))..)))))..)).)))))).... ( -37.70, z-score = -0.98, R) >droEre2.scaffold_4784 20931660 107 + 25762168 UGCACUUGUUGUCCUCCAGCUCGGGCUUAGCUGCUGAGCCUGAGA----------CUCCUGCUGAAGCUAAGGCUGGUCCCUGGAAACAGCUCUAACUGGUGACACAAACAAUAACA .....((((.(((..(((((((((((((((...))))))))))).----------..((.(((........))).))...(((....)))......)))).)))))))......... ( -36.50, z-score = -1.61, R) >droYak2.chr3L 21491100 117 + 24197627 UGCACUUGUUGUCCUCCAGCUCGGGCUUAGCCGCUGAGCCUGAGACUCCCAGUGACUCCCAGUGAAGCUAAAGCUGGACCCUACAACUUCGUCCAACUGGUGACACAAACAAUAACA .....(((((((.....(((((((((((((...))))))))))).))....(((..(((((((..(((....)))((((...........)))).))))).)))))..))))))).. ( -37.40, z-score = -2.35, R) >droSec1.super_0 1469113 107 - 21120651 UGCACUUGUUGUCCUCCAGCUCGGGCUUAGCUGCUGAGCCUGAGA----------CUUAUAGUGAAGCCAAAGCAGGACCCUGGACACAACACUAACUGGUGACACGAACAAUAACA .....((((.(((..(((((((((((((((...))))))))))).----------....(((((..((....))((....))........))))).)))).)))))))......... ( -33.50, z-score = -1.87, R) >droSim1.chr3L 8609896 107 - 22553184 UGCACUUGUUGUCCUCCAGCUCGGGCUUAGCUGCUGAGCCUGAGA----------CUUCUACUGAAGCCAAAGCUGGACCCUGGACACAACACUAACUGGUGACACAAACAAUAACA .....(((((((((((((((((((((((((...))))))))))(.----------((((....)))).)...))))))....))))....((((....)))).....)))))..... ( -37.80, z-score = -3.82, R) >consensus UGCACUUGUUGUCCUCCAGCUCGGGCUUAGCUGCUGAGCCUGAGA__________CUCCUACUGAAGCUAAAGCUGGACCCUGGAAACAACACUAACUGGUGACACAAACAAUAACA ((((((.(((((((.....(((((((((((...))))))))))).....................(((....)))))))...(....)......))).)))).))............ (-22.32 = -22.88 + 0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:13:17 2011