
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,186,671 – 9,186,792 |
| Length | 121 |
| Max. P | 0.877991 |

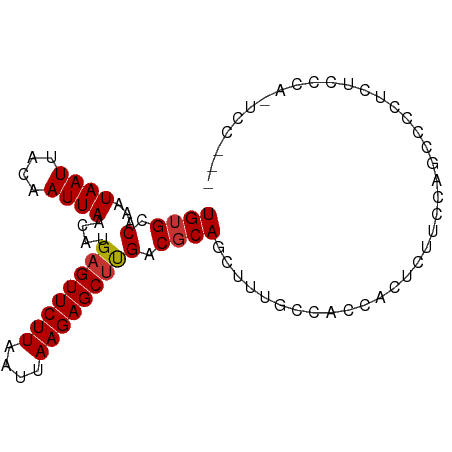
| Location | 9,186,671 – 9,186,761 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 80.48 |
| Shannon entropy | 0.39637 |
| G+C content | 0.41358 |
| Mean single sequence MFE | -12.86 |
| Consensus MFE | -8.75 |
| Energy contribution | -8.76 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.517206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9186671 90 - 24543557 UGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUUGACGCAGCUUUGCCACCACUUUUCCAGCACCUCUCCCAUUCCACU .((((....................((((((((....))))))))...((......)).............))))............... ( -13.00, z-score = -1.44, R) >droSim1.chr3L 8584384 90 - 22553184 UGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUUGACGCAGCUUUGCCACCACUUUUCCAGCCCCUCUCCCAUUCCAGU .(((.((((((((....))))....((((((((....))))))))........)))).)))............................. ( -10.00, z-score = 0.00, R) >droSec1.super_0 1443857 90 - 21120651 UGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUUGACGCAGCUUUGCCACCACUUUUCCAGCCCCUCUCCCAUUCCACU .(((.((((((((....))))....((((((((....))))))))........)))).)))............................. ( -10.00, z-score = -0.41, R) >droYak2.chr3L 21463722 78 + 24197627 UGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUUGACGCAGCUUUGCCACCCCUCCCUCCCUCCACU------------ .(((.((((((((....))))....((((((((....))))))))........)))).))).................------------ ( -10.00, z-score = -1.31, R) >droEre2.scaffold_4784 20901723 86 + 25762168 UGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUUGACGCAGCUUUGCCACCACUUUUCCAGGCCCCAUC----UCCACU ...(((...................((((((((....))))))))...((......)).............)))......----...... ( -12.90, z-score = -0.64, R) >dp4.chrXR_group5 392890 83 + 740492 UGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUCGACGCAGCUUUGGCUCCACUCCUCAAUCCCCGUACAUU------- (((((...................(((((((((....)))))))))..(.(((....))).)..............)))))..------- ( -16.90, z-score = -2.57, R) >droPer1.super_58 66119 83 - 410386 UGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUCGACGCAGCUUUGGCUCCACUCCUCAAUCCCCGUACAUU------- (((((...................(((((((((....)))))))))..(.(((....))).)..............)))))..------- ( -16.90, z-score = -2.57, R) >droWil1.scaffold_181024 379512 87 + 1751709 UGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCACGACUCAGCUUUUGCUUCACACUCUUUGCCACAGUUCUCAGCU--- ((((.((((..................((((((....))))))..((...(((....)))))......)))).))))..........--- ( -13.20, z-score = -0.07, R) >consensus UGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUUGACGCAGCUUUGCCACCACUCUUCCAGCCCCUCUCCCA_UCC___ ((((.(...((((....))))....((((((((....))))))))).))))....................................... ( -8.75 = -8.76 + 0.02)


| Location | 9,186,684 – 9,186,792 |
|---|---|
| Length | 108 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 88.50 |
| Shannon entropy | 0.24311 |
| G+C content | 0.37510 |
| Mean single sequence MFE | -20.75 |
| Consensus MFE | -17.75 |
| Energy contribution | -17.49 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.877991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
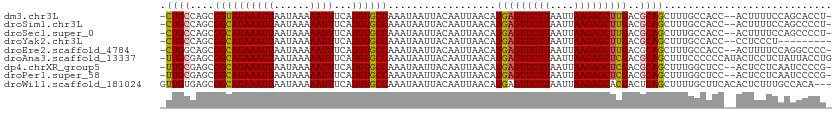
>dm3.chr3L 9186684 108 - 24543557 -CUGCCAGCGGUAUAAAUUAAUAAAAAUUUCAUGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUUGACGCAGCUUUGCCACC--ACUUUUCCAGCACCU- -((((....((((((((((......))))...))))))...................((((((((....))))))))...))))..........--...............- ( -16.70, z-score = -0.26, R) >droSim1.chr3L 8584397 108 - 22553184 -CUGCCAGCGGCAUAAAUUAAUAAAAAUUUCAUGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUUGACGCAGCUUUGCCACC--ACUUUUCCAGCCCCU- -((((....((((((((((......))))...))))))...................((((((((....))))))))...))))..........--...............- ( -18.70, z-score = -1.00, R) >droSec1.super_0 1443870 108 - 21120651 -CUGCCAGCGGCAUAAAUUAAUAAAAAUUUCAUGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUUGACGCAGCUUUGCCACC--ACUUUUCCAGCCCCU- -((((....((((((((((......))))...))))))...................((((((((....))))))))...))))..........--...............- ( -18.70, z-score = -1.00, R) >droYak2.chr3L 21463731 100 + 24197627 -CUGCCAGCGGCAUAAAUUAAUAAAAAUUUCAUGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUUGACGCAGCUUUGCCACC--CCUCCCU--------- -((((....((((((((((......))))...))))))...................((((((((....))))))))...))))..........--.......--------- ( -18.70, z-score = -1.70, R) >droEre2.scaffold_4784 20901732 108 + 25762168 -CUGGCAGCGGCAUAAAUUAAUAAAAAUUUCAUGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUUGACGCAGCUUUGCCACC--ACUUUUCCAGGCCCC- -.(((((((((((((((((......))))...))))))...................((((((((....))))))))......))..)))))..--...............- ( -22.60, z-score = -1.17, R) >droAna3.scaffold_13337 19587838 111 - 23293914 -UUGCGAGCGGCAUAAAUUAAUAAAAAUUUCAUGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUCGACGCAGCUUUCCCCCCAUACUCCUCUAUUACCUG -(((((...((((((((((......))))...))))))..................(((((((((....))))))))).)))))............................ ( -22.40, z-score = -3.37, R) >dp4.chrXR_group5 392896 108 + 740492 -UUGCGAGCGGCAUAAAUUAAUAAAAAUUUCAUGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUCGACGCAGCUUUGGCUCC--ACUCCUCAAUCCCCG- -.((.((((((((((((((......))))...))))))..................(((((((((....)))))))))...........)))))--)..............- ( -23.70, z-score = -2.46, R) >droPer1.super_58 66125 108 - 410386 -UUGCGAGCGGCAUAAAUUAAUAAAAAUUUCAUGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUCGACGCAGCUUUGGCUCC--ACUCCUCAAUCCCCG- -.((.((((((((((((((......))))...))))))..................(((((((((....)))))))))...........)))))--)..............- ( -23.70, z-score = -2.46, R) >droWil1.scaffold_181024 379522 109 + 1751709 GUUGUGAGCGGCAUAAAUUAAUAAAAAUUUCAUGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCACGACUCAGCUUUUGCUUCACACUCUUUGCCACA--- ..((((.((((((((((((......))))...))))))...................((((.......(((((((........)))))))......))))...))))))--- ( -21.52, z-score = -1.39, R) >consensus _CUGCCAGCGGCAUAAAUUAAUAAAAAUUUCAUGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUUGACGCAGCUUUGCCACC__ACUCUUCCAGCCCCU_ .((((....((((((((((......))))...))))))..................(((((((((....)))))))))..))))............................ (-17.75 = -17.49 + -0.26)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:13:14 2011