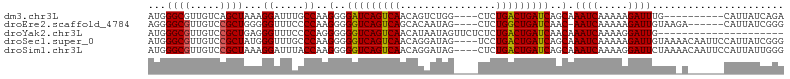
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,186,290 – 9,186,382 |
| Length | 92 |
| Max. P | 0.973224 |

| Location | 9,186,290 – 9,186,382 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 75.15 |
| Shannon entropy | 0.42461 |
| G+C content | 0.46502 |
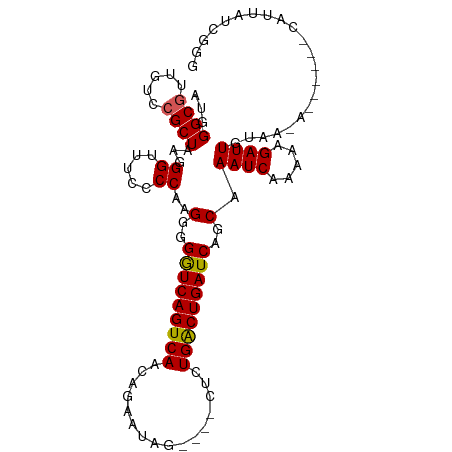
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -15.62 |
| Energy contribution | -15.74 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.973224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
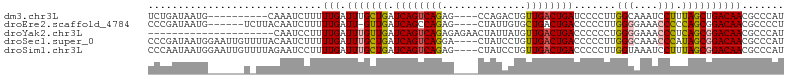
>dm3.chr3L 9186290 92 + 24543557 UCUGAUAAUG----------CAAAUCUUUUUGAUUUGCUGAUCAGUCAGAG----CCAGACUGUUGACUGAUCCCCUUGGCAAAUCCUUUAGCUGACAACGCCCAU .........(----------((((((.....))))))).((((((((((((----.....)).))))))))))...(..((.((....)).))..).......... ( -24.00, z-score = -2.53, R) >droEre2.scaffold_4784 20901354 95 - 25762168 CCCGAUAAUG------UCUUACAAUCUUUUUGAUU-GUUGAUCAGCCAGAG----CUAUUGUGCUGACUGACCCCCUUGGGGAAACCCCCAGCGGACAACGCCCCU ..........------((..((((((.....))))-)).))((((.(((.(----(....)).))).))))..((.((((((....)))))).))........... ( -26.40, z-score = -2.68, R) >droYak2.chr3L 21463348 85 - 24197627 ---------------------CAAUCCUUUUGAUUUGUUGAUCAGUCAGAGAGAACUAUUAUGUUGACUGACCCCCCUGGGGAAACCCUCAGCGGACAACGCCCAU ---------------------........(((.((((((((((((((((.((......))...)))))))).......(((....))))))))))))))....... ( -23.80, z-score = -2.11, R) >droSec1.super_0 1443446 102 + 21120651 CCCGAUAAUGGAAUUGUUUUACAAUCUUUUUGAUUUGCUGAUCAGUCAGGA----CUAUCCUGUUGACUGACCCCCUUGGGCAAACCCAUAGCGGACAACGCCCAU .........((..((((((...((((.....)))).((((.(((((((((.----....))...)))))))......((((....))))))))))))))..))... ( -25.00, z-score = -1.44, R) >droSim1.chr3L 8583973 102 + 22553184 CCCAAUAAUGGAAUUGUUUUAGAAUCCUUUUGAUUUGCUGAUCAGUCAGAG----CUAUCCUGUUGACUGACCCCCUUGGUAAAUCCUUUAGCGGACAACGCCCAU .......((((....................(((((((..(((((((((((----.....)).)))))))).....)..))))))).....(((.....))))))) ( -23.40, z-score = -1.73, R) >consensus CCCGAUAAUG______U_UUACAAUCUUUUUGAUUUGCUGAUCAGUCAGAG____CUAUCCUGUUGACUGACCCCCUUGGGAAAACCCUUAGCGGACAACGCCCAU .............................(((.(((((((.((((((((..............)))))))).......(((....))).))))))))))....... (-15.62 = -15.74 + 0.12)
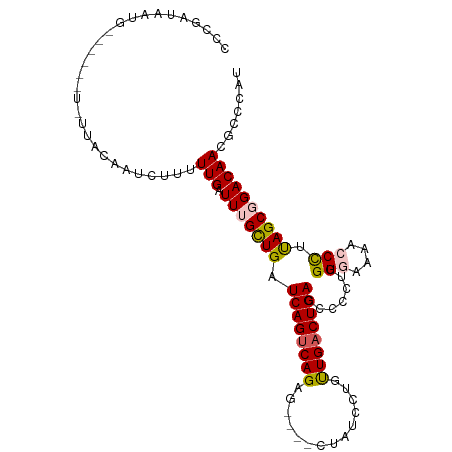
| Location | 9,186,290 – 9,186,382 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 75.15 |
| Shannon entropy | 0.42461 |
| G+C content | 0.46502 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -16.35 |
| Energy contribution | -16.23 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.651270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9186290 92 - 24543557 AUGGGCGUUGUCAGCUAAAGGAUUUGCCAAGGGGAUCAGUCAACAGUCUGG----CUCUGACUGAUCAGCAAAUCAAAAAGAUUUG----------CAUUAUCAGA ...((((..(((........))).)))).....(((((((((..((.....----)).))))))))).(((((((.....))))))----------)......... ( -28.80, z-score = -2.26, R) >droEre2.scaffold_4784 20901354 95 + 25762168 AGGGGCGUUGUCCGCUGGGGGUUUCCCCAAGGGGGUCAGUCAGCACAAUAG----CUCUGGCUGAUCAAC-AAUCAAAAAGAUUGUAAGA------CAUUAUCGGG .....((.(((((.((((((....)))).)).)((((((((((..(....)----..)))))))))).((-((((.....))))))..))------))....)).. ( -32.60, z-score = -2.25, R) >droYak2.chr3L 21463348 85 + 24197627 AUGGGCGUUGUCCGCUGAGGGUUUCCCCAGGGGGGUCAGUCAACAUAAUAGUUCUCUCUGACUGAUCAACAAAUCAAAAGGAUUG--------------------- .(((...((((((.(((.(((...)))))).))(((((((((................))))))))).)))).))).........--------------------- ( -25.19, z-score = -1.03, R) >droSec1.super_0 1443446 102 - 21120651 AUGGGCGUUGUCCGCUAUGGGUUUGCCCAAGGGGGUCAGUCAACAGGAUAG----UCCUGACUGAUCAGCAAAUCAAAAAGAUUGUAAAACAAUUCCAUUAUCGGG (((((.(((((..(((.((((....))))....(((((((((...((....----.)))))))))))))).((((.....)))).....))))))))))....... ( -30.30, z-score = -1.63, R) >droSim1.chr3L 8583973 102 - 22553184 AUGGGCGUUGUCCGCUAAAGGAUUUACCAAGGGGGUCAGUCAACAGGAUAG----CUCUGACUGAUCAGCAAAUCAAAAGGAUUCUAAAACAAUUCCAUUAUUGGG .(((.....((((......))))...))).((((((((((((..((.....----)).)))))))))....((((.....))))..........)))......... ( -22.50, z-score = -0.09, R) >consensus AUGGGCGUUGUCCGCUAAGGGUUUCCCCAAGGGGGUCAGUCAACAGAAUAG____CUCUGACUGAUCAGCAAAUCAAAAAGAUUGUAA_A______CAUUAUCGGG ...((((.....))))...((.....))..(..(((((((((................)))))))))..).((((.....))))...................... (-16.35 = -16.23 + -0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:13:13 2011