| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,184,063 – 9,184,159 |
| Length | 96 |
| Max. P | 0.681527 |

| Location | 9,184,063 – 9,184,159 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 73.49 |
| Shannon entropy | 0.54639 |
| G+C content | 0.45485 |
| Mean single sequence MFE | -18.80 |
| Consensus MFE | -7.79 |
| Energy contribution | -7.72 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.681527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
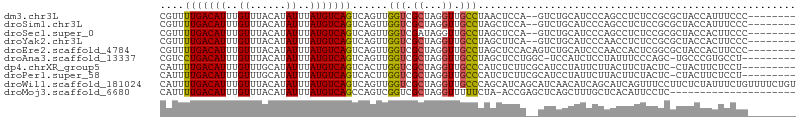
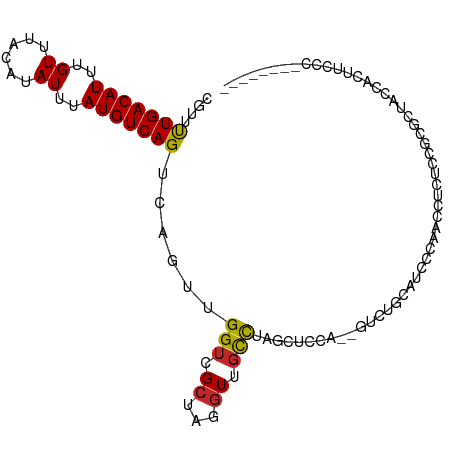
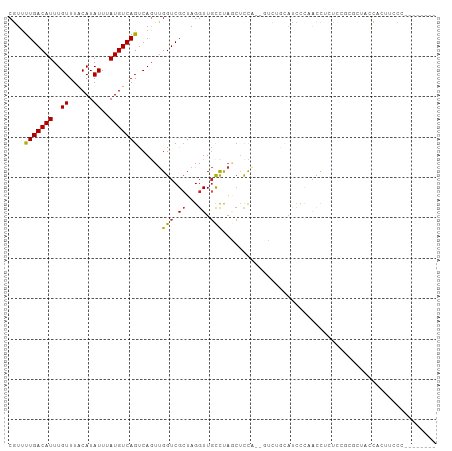
>dm3.chr3L 9184063 96 + 24543557 CGUUUUGACAUUUGUUUACAUAUUUAUGUCAGUCAGUUGGUCGCUAGGUUGCCUAACUCCA--GUCUGCAUCCCAGCCUCUCCGCGCUACCAUUUCCC-------- ....(((((((..((......))..))))))).....((((.((..((.(((...((....--))..))).))..((......)))).))))......-------- ( -18.50, z-score = -1.39, R) >droSim1.chr3L 8581860 96 + 22553184 CGUUUUGACAUUUGUUUACAUAUUUAUGUCAGUCAGUUGGUCGCUAGGUUGCCUAGCUCCA--GUCUGCAUCCCAGCCUCUCCGCGCUACCAUUUCCC-------- .....((((((..((......))..))))))(.(((((((..((((((...)))))).)))--).)))).....(((........)))..........-------- ( -22.60, z-score = -2.54, R) >droSec1.super_0 1441376 96 + 21120651 CGUUUUGACAUUUGUUUACAUAUUUAUGUCAGUCAGUUGGUCGAUAGGUUGCCUAGCUCCA--GUCUGCAUCCCAGCCUCUCCGCGCUACCACUUCCC-------- .....((((((..((......))..))))))(..(((..((.((.((((((....((....--....))....)))))).)).)))))..).......-------- ( -18.60, z-score = -0.82, R) >droYak2.chr3L 21461118 96 - 24197627 CGUUUUGACAUUUGUUUACAUAUUUAUGUCAGUCAGUUGGUCGCUAGGUUGCCUAGCUUCA--GUCUGCAUCCCAACCUCUCCGCGCUACCACUUCCC-------- ....(((((((..((......))..)))))))..(((.((((((.((((((....((....--....))....))))))....)))..))))))....-------- ( -21.10, z-score = -2.16, R) >droEre2.scaffold_4784 20899209 98 - 25762168 CGUUUUGACAUUUGUUUACAUAUUUAUGUCAGUCAGUUGGUCGCUAGGUUGCCUAGCUCCACAGUCUGCAUCCCAACCACUCGGCGCUACCACUUCCC-------- .....((((((..((......))..))))))((((((.(((.((((((...))))))......(....)......)))))).))).............-------- ( -20.20, z-score = -0.82, R) >droAna3.scaffold_13337 19585228 95 + 23293914 CGUCCUGACAUUUGUUUACAUAUUUAUGUCAGUCAGUUGGUCGCUAGGUUGCCUAGCUCCUGGC-UCCAUCUCCUAUUUCCCAGC-UGCCCGUGCCU--------- ....(((((((..((......))..))))))).(((((((..((((((...))))))...((..-..))...........)))))-)).........--------- ( -25.70, z-score = -2.63, R) >dp4.chrXR_group5 390571 96 - 740492 CAUUUUGACAUUUGUUUGCAUAUUUAUGUCAGUCACUUGGUCGCUAGGUUGCCCAUCUCUUCGCAUCCUAUUCUUACUUCUACUC-CUACUUCUCCU--------- ....(((((((..((......))..)))))))............((((.(((..........))).))))...............-...........--------- ( -12.30, z-score = -1.24, R) >droPer1.super_58 63824 96 + 410386 CAUUUUGACAUUUGUUUGCAUAUUUAUGUCAGUCACUUGGUCGCUAGGUUGCCCAUCUCUUCGCAUCCUAUUCUUACUUCUACUC-CUACUUCUCCU--------- ....(((((((..((......))..)))))))............((((.(((..........))).))))...............-...........--------- ( -12.30, z-score = -1.24, R) >droWil1.scaffold_181024 376137 106 - 1751709 CAUUUUGACAUUUGUUUACAUAUUUAUGUCAGUCAGUUGGUCGCUAGGUUGCCCAGCAUCAGCAUCAACAUCAGCAUCAGUUUCCUUCUCUAUUUCUGUUUUCUGU ....(((((((..((......))..)))))))...((((((.(((.(((.((...))))))))))))))..(((...(((...............)))....))). ( -19.16, z-score = -1.53, R) >droMoj3.scaffold_6680 24290398 84 - 24764193 CAUUUUGACAUUUGUUUACAUAUUUAUGUCAGCCAGUCGGUCGCUAGGUUUUUCUA-ACCGAGCUCAGCUUUGCUCACAUUCCUC--------------------- .....((((((..((......))..))))))(((....))).....((((.....)-)))((((........)))).........--------------------- ( -17.50, z-score = -2.25, R) >consensus CGUUUUGACAUUUGUUUACAUAUUUAUGUCAGUCAGUUGGUCGCUAGGUUGCCUAGCUCCA__GUCUGCAUCCCAACCUCUCCGCGCUACCACUUCCC________ ....(((((((..((......))..)))))))......(((.((...)).)))..................................................... ( -7.79 = -7.72 + -0.07)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:13:11 2011