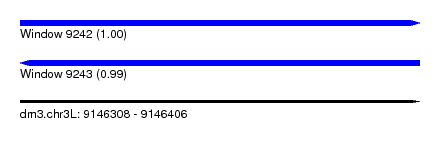
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,146,308 – 9,146,406 |
| Length | 98 |
| Max. P | 0.999796 |

| Location | 9,146,308 – 9,146,406 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 58.10 |
| Shannon entropy | 0.84394 |
| G+C content | 0.39155 |
| Mean single sequence MFE | -27.06 |
| Consensus MFE | -22.62 |
| Energy contribution | -22.45 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.41 |
| SVM RNA-class probability | 0.999796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
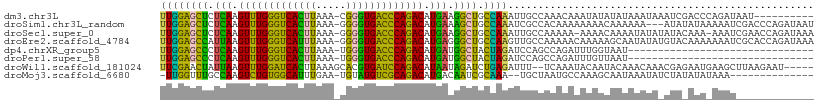
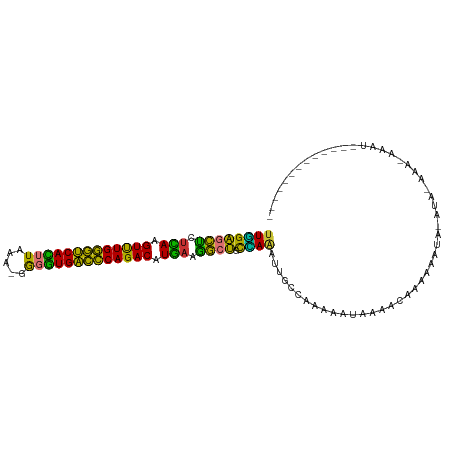
>dm3.chr3L 9146308 98 + 24543557 ----------AUUAUCUGGGUCGAUUUAUUUAUAUAUAUUUGUUUGGCAAUUUGGCAGCCUUCAUGUCUGGGUCACCCG-UUUAAGUGACCCAAACUUGAGAGCUCCAA ----------.........(((((..(((......))).....)))))...((((.(((.((((.((.((((((((...-.....)))))))).)).)))).))))))) ( -29.00, z-score = -2.69, R) >droSim1.chr3L_random 441398 105 + 1049610 AUUAUCUGGGUCGAUUUUUAUAUAU---UUUUUUGUUUUUUUUGUGGCGAUUUGGCAGCUUUCAUGUCUGGGUCACCCC-UUUAAGUGACCCAAACUUGAGAGCUCCAA .........((((........(((.---.....)))........))))...((((.((((((((.((.((((((((...-.....)))))))).)).)))))))))))) ( -33.59, z-score = -4.30, R) >droSec1.super_0 1405126 106 + 21120651 UUUAUCUGGUUCGAUUU-UUUGUAUAUAUAUUUUGUUUU-UUUUUGGCAAUUUGGCAGCCUUCAUGUCUGGGUCACCCC-UUUAAGUGACCCAAACUUGAGAGCUCCAA .................-..............(((((..-.....))))).((((.(((.((((.((.((((((((...-.....)))))))).)).)))).))))))) ( -27.70, z-score = -2.75, R) >droEre2.scaffold_4784 20861808 108 - 25762168 UUUAUCUGGUGCGAUUUUUUUGUACAUAUAUUGCUUUUUGUUUUUGGCAACUUGGCAGCCCUCAUGUCUGGGUCACCCC-UUUAAAUGACCCAAACUUAAUGGCUCCAA ........((((((.....)))))).....(((((..........)))))..(((.((((.....((.(((((((....-......))))))).)).....))))))). ( -29.70, z-score = -2.78, R) >dp4.chrXR_group5 356371 77 - 740492 -------------------------------AUUACCAAAUCUGGCUGGAUCUAGUAGCCAUCAUGUCUGGGUCACCCA-UUUAAGUGACCCAAACUUGAGGGCUCCAA -------------------------------....(((........))).....(.((((.(((.((.((((((((...-.....)))))))).)).))).)))).).. ( -27.30, z-score = -2.96, R) >droPer1.super_58 22198 77 + 410386 -------------------------------AUUAACAAAUCUGGCUGGAUCUAGUAGCCAUCAUGUCUGGGUCACCCA-UUUAAGUGACCCAAACUUGAGGGCUCCAA -------------------------------.............((((....))))((((.(((.((.((((((((...-.....)))))))).)).))).)))).... ( -26.10, z-score = -2.72, R) >droWil1.scaffold_181024 311294 102 - 1751709 -----AUUCUUAAGCUUCAUUCUCGUUUGUUUGUAUUGUAUUUGA--AAAUCUCAGAUCUAUUAUGUCUGGAUCACGUGCUUUAAGUGAUCCAAACUUAAUAGUUCGAA -----.(((..(.((..((..(......)..))....)).)..))--).......((.((((((.((.((((((((.........)))))))).)).)))))).))... ( -22.30, z-score = -2.76, R) >droMoj3.scaffold_6680 24251231 91 - 24764193 --------------UUUAUAUAUAGAUAUUUAUUGCUUUGGCAUUAGCA--UUUGCGAUUGUCAUGUCUGCGACAUACA-UUCAAAUGCCACAGACUUGGCAAACCAA- --------------..................((((....((....)).--...))))((((((.(((((.(.(((...-.....))).).))))).)))))).....- ( -20.80, z-score = -1.59, R) >consensus _____________AUUU_UUU_UAU_UAUUUAUUAUUUUAUUUGUGGCAAUUUGGCAGCCAUCAUGUCUGGGUCACCCA_UUUAAGUGACCCAAACUUGAGAGCUCCAA ....................................................(((.((((((((.((.((((((((.........)))))))).)).))))))))))). (-22.62 = -22.45 + -0.17)


| Location | 9,146,308 – 9,146,406 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 58.10 |
| Shannon entropy | 0.84394 |
| G+C content | 0.39155 |
| Mean single sequence MFE | -32.36 |
| Consensus MFE | -31.60 |
| Energy contribution | -29.15 |
| Covariance contribution | -2.45 |
| Combinations/Pair | 1.75 |
| Mean z-score | -4.79 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.993579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9146308 98 - 24543557 UUGGAGCUCUCAAGUUUGGGUCACUUAAA-CGGGUGACCCAGACAUGAAGGCUGCCAAAUUGCCAAACAAAUAUAUAUAAAUAAAUCGACCCAGAUAAU---------- (((((((..(((.((((((((((((....-..)))))))))))).)))..))).))))..........................(((......)))...---------- ( -34.70, z-score = -5.51, R) >droSim1.chr3L_random 441398 105 - 1049610 UUGGAGCUCUCAAGUUUGGGUCACUUAAA-GGGGUGACCCAGACAUGAAAGCUGCCAAAUCGCCACAAAAAAAACAAAAAA---AUAUAUAAAAAUCGACCCAGAUAAU ((((((((.(((.(((((((((((((...-.))))))))))))).))).)))).)))).......................---..........(((......)))... ( -34.60, z-score = -5.95, R) >droSec1.super_0 1405126 106 - 21120651 UUGGAGCUCUCAAGUUUGGGUCACUUAAA-GGGGUGACCCAGACAUGAAGGCUGCCAAAUUGCCAAAAA-AAAACAAAAUAUAUAUACAAA-AAAUCGAACCAGAUAAA (((((((..(((.(((((((((((((...-.))))))))))))).)))..))).))))...........-.....................-..(((......)))... ( -35.00, z-score = -5.62, R) >droEre2.scaffold_4784 20861808 108 + 25762168 UUGGAGCCAUUAAGUUUGGGUCAUUUAAA-GGGGUGACCCAGACAUGAGGGCUGCCAAGUUGCCAAAAACAAAAAGCAAUAUAUGUACAAAAAAAUCGCACCAGAUAAA ((((((((.(((.(((((((((((((...-.))))))))))))).))).)))).))))(((((............)))))..............(((......)))... ( -34.40, z-score = -3.88, R) >dp4.chrXR_group5 356371 77 + 740492 UUGGAGCCCUCAAGUUUGGGUCACUUAAA-UGGGUGACCCAGACAUGAUGGCUACUAGAUCCAGCCAGAUUUGGUAAU------------------------------- ....((((.(((.(((((((((((((...-.))))))))))))).))).))))((((((((......))))))))...------------------------------- ( -37.60, z-score = -5.78, R) >droPer1.super_58 22198 77 - 410386 UUGGAGCCCUCAAGUUUGGGUCACUUAAA-UGGGUGACCCAGACAUGAUGGCUACUAGAUCCAGCCAGAUUUGUUAAU------------------------------- ....((((.(((.(((((((((((((...-.))))))))))))).))).))))..((((((......)))))).....------------------------------- ( -33.10, z-score = -4.72, R) >droWil1.scaffold_181024 311294 102 + 1751709 UUCGAACUAUUAAGUUUGGAUCACUUAAAGCACGUGAUCCAGACAUAAUAGAUCUGAGAUUU--UCAAAUACAAUACAAACAAACGAGAAUGAAGCUUAAGAAU----- (((((.((((((.(((((((((((.........))))))))))).)))))).))((((((((--((...................))))))....)))).))).----- ( -25.11, z-score = -4.66, R) >droMoj3.scaffold_6680 24251231 91 + 24764193 -UUGGUUUGCCAAGUCUGUGGCAUUUGAA-UGUAUGUCGCAGACAUGACAAUCGCAAA--UGCUAAUGCCAAAGCAAUAAAUAUCUAUAUAUAAA-------------- -((((((((.((.(((((((((((.....-...))))))))))).)).)))..((...--.))....))))).......................-------------- ( -24.40, z-score = -2.22, R) >consensus UUGGAGCUCUCAAGUUUGGGUCACUUAAA_GGGGUGACCCAGACAUGAAGGCUGCCAAAUUGCCAAAAAUAAAACAAAAAAUA_AUA_AAA_AAAU_____________ ((((((((.(((.(((((((((((((.....))))))))))))).))).)))).))))................................................... (-31.60 = -29.15 + -2.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:13:05 2011