| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,136,759 – 9,136,859 |
| Length | 100 |
| Max. P | 0.717579 |

| Location | 9,136,759 – 9,136,859 |
|---|---|
| Length | 100 |
| Sequences | 13 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 83.02 |
| Shannon entropy | 0.38266 |
| G+C content | 0.49692 |
| Mean single sequence MFE | -31.50 |
| Consensus MFE | -21.81 |
| Energy contribution | -22.91 |
| Covariance contribution | 1.10 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.717579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9136759 100 - 24543557 -----------GGCUGU-GCAUCGCUCUCGCCUACUGGUGGGAAAGAUUUAUCGGCACCGUCUUCAACCAUGGGGGUUGCAGCGGUAACAAUUAUUUGUGCCAUACCUAAGU -----------((..((-(.(((..(((((((....)))))))..))).))).(((((((.((.(((((.....))))).)))))).((((....)))))))...))..... ( -33.80, z-score = -1.59, R) >droSim1.chr3L 8544680 100 - 22553184 -----------GGCUGC-GCAUCGCUCUCGCCUACUGGUGGGAAAGAUUUAUCGGCACUGUCUUCAACCAUGGGGGUUGCAGCGGUAACAAUUAUUUGUGCCAUACCUAAGU -----------((....-..(((..(((((((....)))))))..))).....(((((((.((.(((((.....))))).)))))).((((....)))))))...))..... ( -31.00, z-score = -0.82, R) >droSec1.super_0 1395754 100 - 21120651 -----------GGCUGC-GCAUCGCUCUCGCCUACUGGUGGGAAAGAUUUAUCGGCACCGUCUUCAACCAUGGGGGUUGCAGCGGUAACAAUUAUUUGUGCCAUACCUAAGU -----------((....-..(((..(((((((....)))))))..))).....(((((((.((.(((((.....))))).)))))).((((....)))))))...))..... ( -33.20, z-score = -1.30, R) >droYak2.chr3L 21413334 100 + 24197627 -----------GGCUGC-GCAUCGCUCUCGCCUACUGGUGGGAAAGAUUUAUCGGCACCGUCUUCAACCAUGGGGGUUGCAGCGGUAACAAUUAUUUGUGCCAUACCUAAGU -----------((....-..(((..(((((((....)))))))..))).....(((((((.((.(((((.....))))).)))))).((((....)))))))...))..... ( -33.20, z-score = -1.30, R) >droEre2.scaffold_4784 20852203 100 + 25762168 -----------GGCUGC-GCAUCGCUCUCGCCUACUGGUGGGAAAGAUUUAUCGGCACCGUCUUCAACCAUGGGGGUUGCAGCGGUAACAAUUAUUUGUGCCAUACCUAAGU -----------((....-..(((..(((((((....)))))))..))).....(((((((.((.(((((.....))))).)))))).((((....)))))))...))..... ( -33.20, z-score = -1.30, R) >droAna3.scaffold_13337 19534869 111 - 23293914 UGUCCUUAUCGGACUGG-GAAUCGCUCUCUCCUACUGGUGGGAAAGAUUUAUCGGCACCGUCUUCAACCAUGGGGGUUGCAGCGGUAACUAUUAUUUGUGCCAUACCUAAGU .((((.....))))(((-(((((..((((.((....)).))))..))))....(((((((.((.(((((.....))))).)))))).((........)))))...))))... ( -36.40, z-score = -1.81, R) >dp4.chrXR_group5 345975 100 + 740492 -----------UGCUCU-CUUUCGCUCUCUCCUACUGGUGGGAAAGAUUUAUCGGCACCGUCUUCAACCAUGGGGGUUGCAGCGGUAACAAUUAUUUGUGCCAUACCUAAGU -----------.((...-.....))(((.(((((....))))).)))......(((((((.((.(((((.....))))).)))))).((((....))))))).......... ( -28.40, z-score = -1.29, R) >droPer1.super_58 11628 100 - 410386 -----------UGCUCU-CUUUCGCUCUCUCCUACUGGUGGGAAAGAUUUAUCGGCACCGUCUUCAACCAUGGGGGUUGCAGCGGUAACAAUUAUUUGUGCCAUACCUAAGU -----------.((...-.....))(((.(((((....))))).)))......(((((((.((.(((((.....))))).)))))).((((....))))))).......... ( -28.40, z-score = -1.29, R) >droWil1.scaffold_181024 294640 100 + 1751709 -----------UGCUGU-UCCUCCUUCUCUCCUACUGGUGGGAAAGAUUUAUCUGCACCGUCUUCAACCAUUGGGGUUGCAGCGGUAACUAUUAUUUGUGCCAUACCUAAAU -----------.(((((-..(((((((((.((....)).)))))(((....)))..................))))..)))))((((.(........))))).......... ( -25.90, z-score = -1.42, R) >droVir3.scaffold_13049 711480 95 - 25233164 -----------------AUUGUCCCUUUCUCCUACUGGUGGGAAAGAUUUAUCGGCACCGUCUUCAACCAUUGGGGUUGCAGCGGUAACAAUUAUUUGUGCCAUACCUAAAU -----------------.......((((((((.....).))))))).......(((((((.((.(((((.....))))).)))))).((((....))))))).......... ( -27.30, z-score = -1.69, R) >droMoj3.scaffold_6680 24240113 100 + 24764193 ------------UCUGUAUUGUCGCUCUCUCCUACUGGUGGGAAAGAUUUAUCGGCACCGUCUUCAACCAUUGGGGUUGCAGCGGUAACAAUUAUUUGUGCCAUACCUAAGU ------------...((((.(((..((((.((....)).))))..))).....(((((((.((.(((((.....))))).)))))).((((....)))))))))))...... ( -29.70, z-score = -1.91, R) >droGri2.scaffold_15110 1945084 100 - 24565398 ------------GAUGAAUUGUCGCUCUCUCCUACUGGUGGGAAAGAUUUAUCGGCACCGUCUUCAACCAUUGGGGUUGCAGCGGUAACAAUUAUUUGUGCCAUACCUAAGU ------------((((((((.....((((.((....)).))))..))))))))(((((((.((.(((((.....))))).)))))).((((....))))))).......... ( -33.10, z-score = -2.78, R) >anoGam1.chr2L 11246776 103 + 48795086 --GACUCGGCGUCGUCCGCUGUCG-GCCCUUCUUCCGUCGGCAGGAACUUCUUGUCCGGCUCAUCGUGCAUCGGAGUAGCGGCCGUGACGAUGAUCUGCGCAAAGU------ --.(((..((((.(..((.(((((-(((((..(((((.(((((((.....)))).)))((.......))..))))).)).))))..)))).))..).))))..)))------ ( -35.90, z-score = 0.50, R) >consensus ___________GGCUGC_GCAUCGCUCUCUCCUACUGGUGGGAAAGAUUUAUCGGCACCGUCUUCAACCAUGGGGGUUGCAGCGGUAACAAUUAUUUGUGCCAUACCUAAGU ....................(((..((((.((....)).))))..))).....(((((((.((.(((((.....))))).)))))).((((....))))))).......... (-21.81 = -22.91 + 1.10)

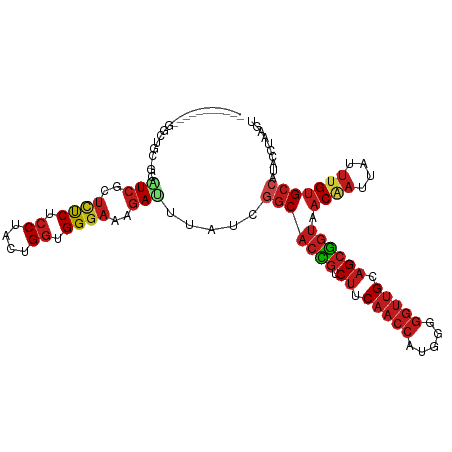

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:13:04 2011