| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,117,319 – 9,117,410 |
| Length | 91 |
| Max. P | 0.967111 |

| Location | 9,117,319 – 9,117,410 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 68.39 |
| Shannon entropy | 0.61068 |
| G+C content | 0.47680 |
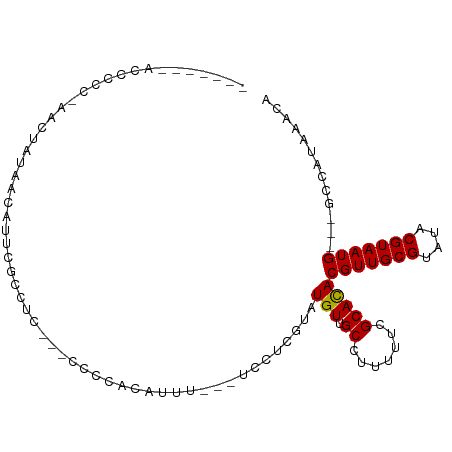
| Mean single sequence MFE | -17.28 |
| Consensus MFE | -7.65 |
| Energy contribution | -7.54 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
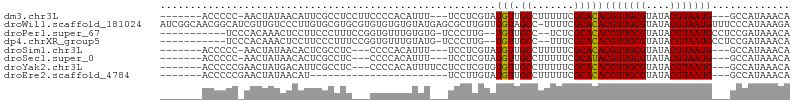
>dm3.chr3L 9117319 91 - 24543557 -------ACCCCC-AACUAUAACAUUCGCCUCCUUCCCCACAUUU---UCCUCGUAUGUUGCCUUUUUCGCACACGUUGCGUAUACGUAAUG---GCCAUAAACA -------......-.............(((...............---.........(((((.......))).))((((((....)))))))---))........ ( -10.90, z-score = -1.06, R) >droWil1.scaffold_181024 269201 104 + 1751709 AUCGGCAACGGCAUCGUUGUCCCUUGUGCGUGCGUGUGUGUGUAUGAGCGCUUGUUUGUAGCC-UUUUCGCACACGUUGCGUAUACGUAAUGUUUCCCAUAAAGA ...(((((((....)))))))...((((((((((((((((.....(((.(((.......))))-))..)))))))).)))))))).................... ( -32.90, z-score = -1.31, R) >droPer1.super_67 138687 89 + 321716 -----------UCCCACAAACUCCUUCCCUUUCCGGUGUUUGUGUG-UCCCUUG--UGUUGCC--UCUCGCACACGUUGCGUAUACGUAAUGCCUCCGAUAAACA -----------...(((((((.((..........)).)))))))((-((...((--(((....--....)))))(((((((....))))))).....)))).... ( -22.00, z-score = -3.87, R) >dp4.chrXR_group5 550465 89 - 740492 -----------UCCCACAAACUCCUUCCCUUUCCGGUGUUUGUAUG-UCCCUUG--UGUUGCC--UUUCGCACACGUUGCGUAUACGUAAUGCCUCCGAUAAACA -----------....((((((.((..........)).)))))).((-((...((--(((....--....)))))(((((((....))))))).....)))).... ( -19.60, z-score = -3.30, R) >droSim1.chr3L 8529099 88 - 22553184 -------ACCCCC-AACUAUAACACUCGCCUC---CCCCACAUUU---UCCUCGUAUGUUGCCUUUUUCGCACACGUUGCGUAUACGUAAUG---GCCAUAAACA -------......-.............(((..---..........---.........(((((.......))).))((((((....)))))))---))........ ( -10.90, z-score = -0.89, R) >droSec1.super_0 1380127 88 - 21120651 -------ACCCCC-AACUAUAACACUCGCCUC---CCCCACAUUU---UCCUCGUAUGUUGCCUUUUUCGCAUACGUUGCGUAUACGUAAUG---GCCAUAAACA -------......-.............(((..---..........---.....((((((..........))))))((((((....)))))))---))........ ( -14.20, z-score = -2.38, R) >droYak2.chr3L 21397322 92 + 24197627 -------ACCCCCGAACUAUGACAUUCGCCUC---CCCCACAUUUUCCUCCUCGUGUGUUGCCUUUUUCGCACACGUUGCGUAUACGUAAUG---GCCAUAAACA -------....................(((..---..................((((((..........))))))((((((....)))))))---))........ ( -16.10, z-score = -1.67, R) >droEre2.scaffold_4784 20834764 72 + 25762168 -------ACCCCCGAACUAUAACAU-----------------------UCCUUGUAUGUUGCCUUUUUCGCACACGUUGCGUAUACGUAAUG---GCCAUAAACA -------.....((((...((((((-----------------------.......)))))).....))))....(((((((....)))))))---.......... ( -11.60, z-score = -0.67, R) >consensus _______ACCCCC_AACUAUAACAUUCGCCUC___CCCCACAUUU___UCCUCGUAUGUUGCCUUUUUCGCACACGUUGCGUAUACGUAAUG___GCCAUAAACA ........................................................(((.((.......)))))(((((((....)))))))............. ( -7.65 = -7.54 + -0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:13:02 2011