
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,644,264 – 4,644,415 |
| Length | 151 |
| Max. P | 0.958811 |

| Location | 4,644,264 – 4,644,415 |
|---|---|
| Length | 151 |
| Sequences | 6 |
| Columns | 153 |
| Reading direction | forward |
| Mean pairwise identity | 67.81 |
| Shannon entropy | 0.64139 |
| G+C content | 0.52559 |
| Mean single sequence MFE | -46.73 |
| Consensus MFE | -19.68 |
| Energy contribution | -20.29 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.925574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
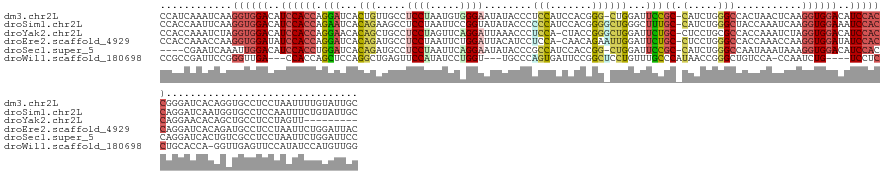
>dm3.chr2L 4644264 151 + 23011544 CCAUCAAAUCAAGGUGGACAUCCACCAGGAUCACUGUUGCCUCCUAAUGUGGGAAUAUACCCUCCAUCCACGGG-CUGGAUUCCGC-CAUCUGGGCCACUAACUCAAGGUGGACAUCCACCGGGAUCACAGGUGCCUCCUAAUUUUGUAUUGC .((.((((....(((((....)))))((((.((((...((((....(((.(((......)))..)))....)))-)((((((((((-(.....)))...........(((((....))))))))))).))))))..))))...))))...)). ( -49.30, z-score = -0.89, R) >droSim1.chr2L 4518132 152 + 22036055 CCACCAAUUCAAGGUGGACAUCCACCAGAAUCACAGAAGCCUCCUAAUUCCGGUAUAUACCCCCCAUCCACGGGGCUGGGCUUUGC-CAUCUGGGCUACCAAAUCAAGGUGGAAAUCCACCAGGAUCAAUGGUGCCUCCAAUUUCUGUAUUGC .(((((..((..((((((..((((((.........(((((((.........(((....)))((((......))))..)))))))((-(.....)))...........))))))..))))))..))....)))))....((((......)))). ( -51.10, z-score = -1.91, R) >droYak2.chr2L 4658385 142 + 22324452 CCACCAAAUCUAGGUGGACAUCCACCAGGAACACAGCUGCCUCCUAGUUCAGGAUUAAACCCUCCA-CUACCGGGCUGGAUUCUGC-CUCCUGCGCCACCAAAUCUAGGUGGACAUCCACCAGGAACACAGCUGCCUCCUAGUU--------- ............(((((....)))))((((...((((((..((((.((.(((((.....(((....-.....)))..((......)-)))))).))...........(((((....)))))))))...))))))..))))....--------- ( -47.20, z-score = -2.56, R) >droEre2.scaffold_4929 4724246 151 + 26641161 CCACCAAACCAAGGUGGAUAUCCACCAGGAUCACAGAUGCCUCCUAAUUCUGGAUUACAUCCUCCA-CAACAGAAUUGGAUUCUGC-CUCCUGGGCCACCAAACCAAGGUGGAUAUCCACCAGGAUCACAGAUGCCUCCUAAUUCUGGAUUAC ............((((((((((((((((((...((((....(((.(((((((..............-...)))))))))).)))).-.))))((....)).......))))))))))))))..((((.((((...........)))))))).. ( -53.83, z-score = -4.31, R) >droSec1.super_5 2728994 147 + 5866729 ----CGAAUCAAAUUGGACAUCCACCUGGAUCACAGAUGCCUCCUAAUUCAGGAAUAUACCCGCCAUCCACCGG-CUGGAUUCCGC-CAUCUGGGCCAAUAAAUAAAGGUGGACAUCCACCAGGAUCACUGUCGCCUCCUAAUUCUGGAUUCC ----.(((((....((((..(((((((((.((.((((((.(((((.....)))).....((.(((.......))-).)).....).-)))))))))).........)))))))..)))).((((((...............))))))))))). ( -42.76, z-score = -1.38, R) >droWil1.scaffold_180698 4445315 141 - 11422946 CCGCCGAUUCCGGGUUGA---CCACCAGCUCCAGGCUGAGUUCCAUAUCCUGGU---UGCCCAGUGAUUCCGGCUCCUGUUUGCCCAUAACCGGGCUGUCCA-CCAAUCUG----UCCUCCUGCACCA-GGUUGAGUUCCAUAUCCAUGUUGG ...(((((..(((((((.---..((((((.....)))).)).........(((.---.((((.((......(((........)))....)).))))...)))-.)))))))----((..((((...))-))..)).............))))) ( -36.20, z-score = 1.28, R) >consensus CCACCAAAUCAAGGUGGACAUCCACCAGGAUCACAGAUGCCUCCUAAUUCAGGAAUAUACCCUCCAUCCACGGG_CUGGAUUCCGC_CAUCUGGGCCACCAAAUCAAGGUGGACAUCCACCAGGAUCACAGGUGCCUCCUAAUUCUGUAUUGC ............((((((..((((((.(((...(((...(((.........((.......)).........))).)))...))).......(((....)))......))))))..))))))................................ (-19.68 = -20.29 + 0.60)
| Location | 4,644,264 – 4,644,415 |
|---|---|
| Length | 151 |
| Sequences | 6 |
| Columns | 153 |
| Reading direction | reverse |
| Mean pairwise identity | 67.81 |
| Shannon entropy | 0.64139 |
| G+C content | 0.52559 |
| Mean single sequence MFE | -56.17 |
| Consensus MFE | -25.23 |
| Energy contribution | -25.73 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.958811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
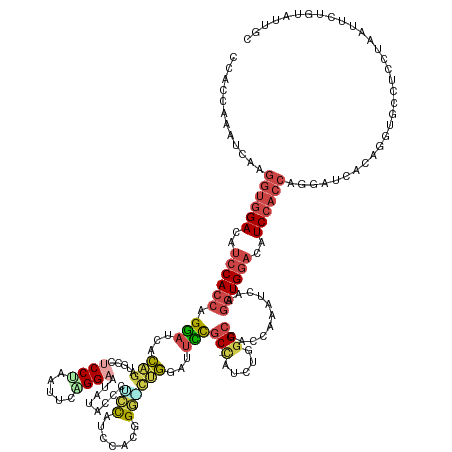
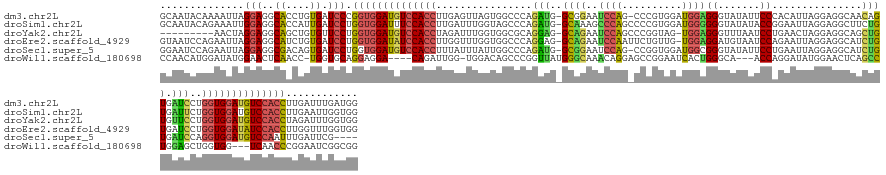
>dm3.chr2L 4644264 151 - 23011544 GCAAUACAAAAUUAGGAGGCACCUGUGAUCCCGGUGGAUGUCCACCUUGAGUUAGUGGCCCAGAUG-GCGGAAUCCAG-CCCGUGGAUGGAGGGUAUAUUCCCACAUUAGGAGGCAACAGUGAUCCUGGUGGAUGUCCACCUUGAUUUGAUGG ........(((((((((..((....)).))).((((((((((((((..(..(((.(((((.....)-))........(-((....((((..(((......))).))))....)))..)).)))..).)))))))))))))).))))))..... ( -53.80, z-score = -0.88, R) >droSim1.chr2L 4518132 152 - 22036055 GCAAUACAGAAAUUGGAGGCACCAUUGAUCCUGGUGGAUUUCCACCUUGAUUUGGUAGCCCAGAUG-GCAAAGCCCAGCCCCGUGGAUGGGGGGUAUAUACCGGAAUUAGGAGGCUUCUGUGAUUCUGGUGGAUGUCCACCUUGAAUUGGUGG ((....(((...)))...)).((((..(((..(((((((.((((((...((((((....)))))).-((((((((..(((((.(....).))))).....((.......)).))))).)))......)))))).)))))))..).))..)))) ( -56.40, z-score = -1.19, R) >droYak2.chr2L 4658385 142 - 22324452 ---------AACUAGGAGGCAGCUGUGUUCCUGGUGGAUGUCCACCUAGAUUUGGUGGCGCAGGAG-GCAGAAUCCAGCCCGGUAG-UGGAGGGUUUAAUCCUGAACUAGGAGGCAGCUGUGUUCCUGGUGGAUGUCCACCUAGAUUUGGUGG ---------.((((((((((((((((.((((((((....(.(((((.......)))))).(((((.-...((((((..(((....)-.)).))))))..))))).)))))))))))))))).))))))))......(((((.......))))) ( -63.00, z-score = -3.16, R) >droEre2.scaffold_4929 4724246 151 - 26641161 GUAAUCCAGAAUUAGGAGGCAUCUGUGAUCCUGGUGGAUAUCCACCUUGGUUUGGUGGCCCAGGAG-GCAGAAUCCAAUUCUGUUG-UGGAGGAUGUAAUCCAGAAUUAGGAGGCAUCUGUGAUCCUGGUGGAUAUCCACCUUGGUUUGGUGG ....(((.......)))..((((...((((..((((((((((((((..((((....)))).((((.-(((((..((..(((((((.-((((........)))).)).)))))))..)))))..))))))))))))))))))..)))).)))). ( -59.80, z-score = -2.48, R) >droSec1.super_5 2728994 147 - 5866729 GGAAUCCAGAAUUAGGAGGCGACAGUGAUCCUGGUGGAUGUCCACCUUUAUUUAUUGGCCCAGAUG-GCGGAAUCCAG-CCGGUGGAUGGCGGGUAUAUUCCUGAAUUAGGAGGCAUCUGUGAUCCAGGUGGAUGUCCAAUUUGAUUCG---- .(((((.((((((((((.((....))..)))))))((((((((((((......((((((...(((.-.....)))..)-)))))((((.((((((...((((((...))))))..)))))).))))))))))))))))..)))))))).---- ( -59.30, z-score = -2.90, R) >droWil1.scaffold_180698 4445315 141 + 11422946 CCAACAUGGAUAUGGAACUCAACC-UGGUGCAGGAGGA----CAGAUUGG-UGGACAGCCCGGUUAUGGGCAAACAGGAGCCGGAAUCACUGGGCA---ACCAGGAUAUGGAACUCAGCCUGGAGCUGGUGG---UCAACCCGGAAUCGGCGG ((..(((.(((.((...((...((-((...))))))..----)).))).)-))....(((((....))))).....)).(((((.(((.((((...---.))))))).(((.((.((((.....))))))..---)))........))))).. ( -44.70, z-score = 0.74, R) >consensus GCAAUACAGAAUUAGGAGGCAACUGUGAUCCUGGUGGAUGUCCACCUUGAUUUGGUGGCCCAGAUG_GCAGAAUCCAG_CCCGUGGAUGGAGGGUAUAUUCCAGAAUUAGGAGGCAUCUGUGAUCCUGGUGGAUGUCCACCUUGAUUUGGUGG ..............(((..((....)).))).((((((((((((((...............(((...((((..((((..........))))((.......))...............))))...)))))))))))))))))............ (-25.23 = -25.73 + 0.50)
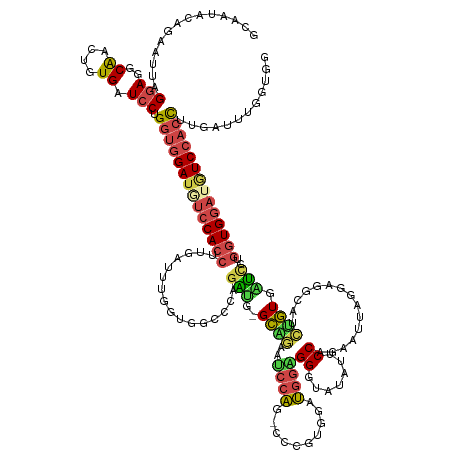
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:17:06 2011