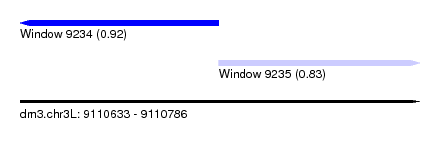
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,110,633 – 9,110,786 |
| Length | 153 |
| Max. P | 0.922103 |

| Location | 9,110,633 – 9,110,709 |
|---|---|
| Length | 76 |
| Sequences | 4 |
| Columns | 79 |
| Reading direction | reverse |
| Mean pairwise identity | 90.32 |
| Shannon entropy | 0.15094 |
| G+C content | 0.54063 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -23.30 |
| Energy contribution | -23.93 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.922103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
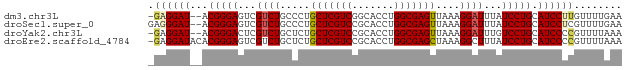
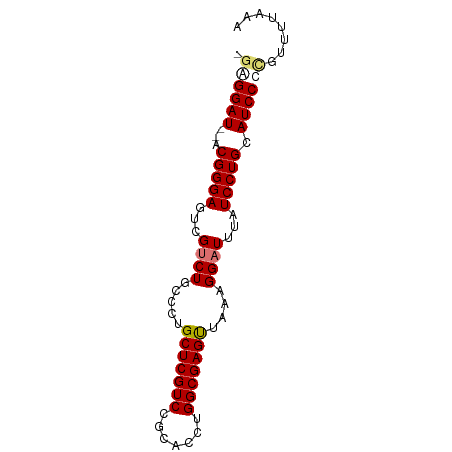
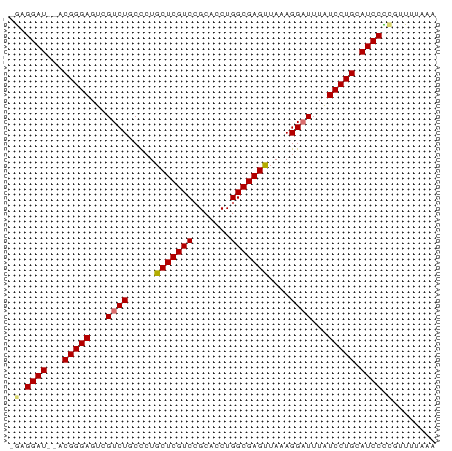
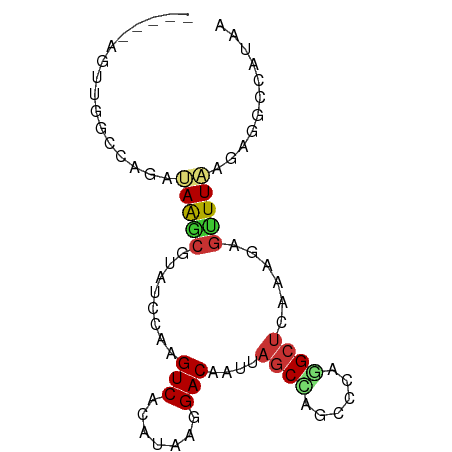
>dm3.chr3L 9110633 76 - 24543557 -GAGGAU--ACGGGAGUCGUCUGCCCUGCUCGUCGGCACCUGGCGAGUUAAAGGAUUUAUCCUGCAUCCUUGUUUUGAA -((((((--.(((((...((((.....(((((((((...)))))))))....))))...))))).))))))........ ( -30.80, z-score = -3.16, R) >droSec1.super_0 1373464 77 - 21120651 GAGGGAU--ACGGGAGUCGUCUGCCCUGCUCGUCCGCACCUGGCGAGUUAAAGGAUUUAUCCUGCAUCCUCGUUUUGAA ((((.((--.(((((...((((.....(((((((.......)))))))....))))...))))).))))))........ ( -26.70, z-score = -1.45, R) >droYak2.chr3L 21390277 76 + 24197627 -GAGGAU--ACGGGACUCGUCUGCUCUGCUCGUCCGCACCUGGCGAGUUAAAGGAUUUGUCCUGCAUCCCCGUUUUAAA -(.((((--.((((((..((((.....(((((((.......)))))))....))))..)))))).)))).)........ ( -29.00, z-score = -3.04, R) >droEre2.scaffold_4784 20827999 78 + 25762168 -GAGGAUACACGGGAGUCGUCUGCUCUGCUCGUCCGCACCUGGCGAGCUAAAGGCUUUAUCCUGCAUCCCCGUUUUAAA -(.((((...(((((...((((.....(((((((.......)))))))...))))....))))).)))).)........ ( -24.80, z-score = -1.22, R) >consensus _GAGGAU__ACGGGAGUCGUCUGCCCUGCUCGUCCGCACCUGGCGAGUUAAAGGAUUUAUCCUGCAUCCCCGUUUUAAA .((((((...(((((...((((.....(((((((.......)))))))....))))...))))).))))))........ (-23.30 = -23.93 + 0.63)
| Location | 9,110,709 – 9,110,786 |
|---|---|
| Length | 77 |
| Sequences | 8 |
| Columns | 82 |
| Reading direction | forward |
| Mean pairwise identity | 70.29 |
| Shannon entropy | 0.60464 |
| G+C content | 0.41225 |
| Mean single sequence MFE | -16.15 |
| Consensus MFE | -6.29 |
| Energy contribution | -6.57 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.829383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9110709 77 + 24543557 -----AGUUGGCCAGAUAAGCGUAUCCAAGUCACAUAAGGACAAUUAGCCAGCUCAGGCUCAAAGAGUUUAAGAGGCCAUAA -----...(((((...(((((........(((.......)))....((((......))))......)))))...)))))... ( -19.00, z-score = -2.26, R) >droEre2.scaffold_4784 20828077 77 - 25762168 -----AGUUGGCCAGAUAAGCGUAUCCAAGUCACAUAAGGACAAUUAGCCAGCCCAGGCUCAAAGAGUUUAAGAGGCCAUAA -----...(((((...(((((........(((.......)))....((((......))))......)))))...)))))... ( -19.00, z-score = -2.43, R) >droYak2.chr3L 21390353 77 - 24197627 -----AGUUGGCCAGAUAAGCGUAUCCAAGUCACAUAAGGACAAUUAGCCAGCCCAGGCUCAAAGAGUUUAAGAGGCCAUAA -----...(((((...(((((........(((.......)))....((((......))))......)))))...)))))... ( -19.00, z-score = -2.43, R) >droSec1.super_0 1373541 77 + 21120651 -----AGUUGGCCAGAUAAGCGUAUCCAAGUCACAUAAGGACAAUUAGCCAGCUCAGGCUCAAAGAGUUUAAGAGGCCAUAA -----...(((((...(((((........(((.......)))....((((......))))......)))))...)))))... ( -19.00, z-score = -2.26, R) >droAna3.scaffold_13337 19514041 79 + 23293914 ---GCUGGGUCAGAGAUAACCGCAUCCAAGUCACAUAAGGACAAUUAGCUCGCCCAGGCUCAAAGAGUUUAAGAGGACAUAA ---.((((((.((.(((......)))...(((.......)))......)).))))))((((...)))).............. ( -14.60, z-score = 0.08, R) >droVir3.scaffold_13049 23630068 70 - 25233164 GUUGGCAACAAGCUGAUAAGCGUAUGUAAGUCACAUAAGGACAAUUAGCCAA---AAGCUCAAGAGGCUUUAA--------- ..((((.(((.(((....)))...)))..(((.......))).....))))(---(((((.....))))))..--------- ( -16.90, z-score = -1.76, R) >droMoj3.scaffold_6654 2218435 65 + 2564135 -----AAUUUGUCUGAUAAGCGUAUGUAAGUCACAUAAGGACAAUUAGCCAA---AAGCUCAAGAGGCUUUAA--------- -----...((((((........(((((.....))))).)))))).......(---(((((.....))))))..--------- ( -11.30, z-score = -0.59, R) >droWil1.scaffold_181024 261567 67 - 1751709 ---------------ACAGAUAUAUGUAAGUCACAUAAGGACAAUUAGCUCGUAGAAGAAAAAAGUUUUUGAAAGAGUUAAA ---------------(((......)))..(((.......)))..(((((((...(((((......)))))....))))))). ( -10.40, z-score = -1.98, R) >consensus _____AGUUGGCCAGAUAAGCGUAUCCAAGUCACAUAAGGACAAUUAGCCAGCCCAGGCUCAAAGAGUUUAAGAGGCCAUAA ................(((((........(((.......)))....((((......))))......)))))........... ( -6.29 = -6.57 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:12:59 2011