| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,077,330 – 9,077,421 |
| Length | 91 |
| Max. P | 0.912997 |

| Location | 9,077,330 – 9,077,421 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 54.18 |
| Shannon entropy | 0.66041 |
| G+C content | 0.53923 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -14.78 |
| Energy contribution | -14.90 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.27 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.653299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
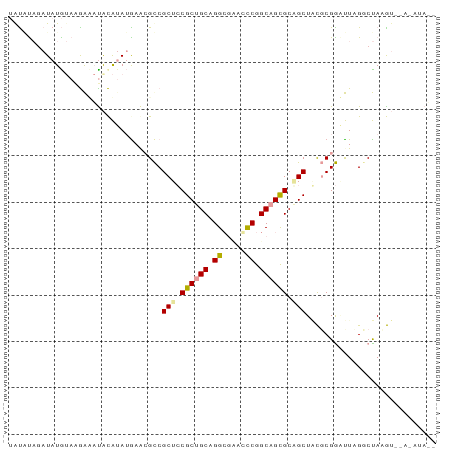
>dm3.chr3L 9077330 91 + 24543557 AGCCUAGAUAUAUAAGAUAUACAUAAGAACGCCGCUCCGCUGCUGGCGUACCCGGCAGCGCAGCUACGCGGAUUAGCCUAAGUCCAAAUAU- ..............................((.(((.(((((((((.....))))))))).)))...))(((((......)))))......- ( -27.20, z-score = -1.11, R) >droSim1.chr3h_random 1352253 82 + 1452968 UAUAUAUAUAUGUAGGAAACGCAAAUAAGCGUCGCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCCUCGACUUAGGCUAAG---------- ..........((.((((.((((......)))).((.(((((((.(((...))).))))))).))))))))............---------- ( -30.10, z-score = -1.54, R) >droVir3.scaffold_13036 524545 90 - 1135500 UAUGCUGAUAUG-ACAGCCUAUGUAUGAA-GCCGCUCCGCCGCGGGGGCGUUCGGCAGUGCGGCUGUGCGGGCUUGGCCAUAUGGAGAUAGA ...((((.....-.))))((((((.(.((-(((((.(.((((((..(.(....).)..)))))).).)).))))).)..))))))....... ( -33.00, z-score = 0.50, R) >consensus UAUAUAGAUAUGUAAGAAAUACAUAUGAACGCCGCUCCGCUGCAGGCGAACCCGGCAGCGCAGCUACGCGGAUUAGGCUAAGU__A_AUA__ .................................(((.((((((.((.....)).)))))).)))............................ (-14.78 = -14.90 + 0.12)


| Location | 9,077,330 – 9,077,421 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 54.18 |
| Shannon entropy | 0.66041 |
| G+C content | 0.53923 |
| Mean single sequence MFE | -31.38 |
| Consensus MFE | -12.80 |
| Energy contribution | -13.80 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.912997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
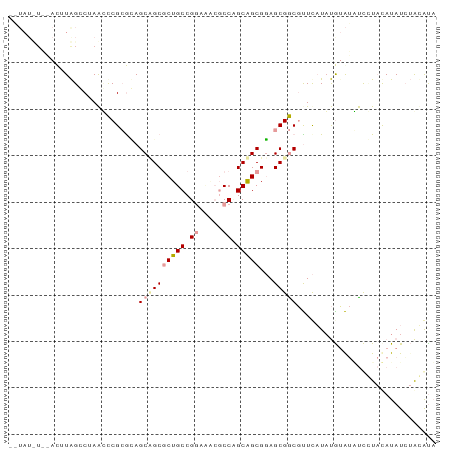
>dm3.chr3L 9077330 91 - 24543557 -AUAUUUGGACUUAGGCUAAUCCGCGUAGCUGCGCUGCCGGGUACGCCAGCAGCGGAGCGGCGUUCUUAUGUAUAUCUUAUAUAUCUAGGCU -......((.((((((.......((((.(((.((((((.((.....)).)))))).))).))))...((((((.....)))))))))))))) ( -31.40, z-score = -0.65, R) >droSim1.chr3h_random 1352253 82 - 1452968 ----------CUUAGCCUAAGUCGAGGAGCAGCGCUGCCGGCAUAGCCUGCAGCGCAGCGACGCUUAUUUGCGUUUCCUACAUAUAUAUAUA ----------..............((((((.(((((((.(((...))).))))))).))(((((......)))))))))............. ( -31.70, z-score = -2.55, R) >droVir3.scaffold_13036 524545 90 + 1135500 UCUAUCUCCAUAUGGCCAAGCCCGCACAGCCGCACUGCCGAACGCCCCCGCGGCGGAGCGGC-UUCAUACAUAGGCUGU-CAUAUCAGCAUA .........(((((((..((((.....((((((.((((((..........)))))).)))))-).........))))))-)))))....... ( -31.04, z-score = -1.49, R) >consensus __UAU_U__ACUUAGCCUAACCCGCGCAGCAGCGCUGCCGGAAACGCCAGCAGCGGAGCGGCGUUCAUAUGUAUAUCCUACAUAUCUACAUA ............................((((((((((.((.....)).)))))...))))).............................. (-12.80 = -13.80 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:12:56 2011