
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,067,876 – 9,067,982 |
| Length | 106 |
| Max. P | 0.879923 |

| Location | 9,067,876 – 9,067,982 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 63.70 |
| Shannon entropy | 0.70891 |
| G+C content | 0.38805 |
| Mean single sequence MFE | -24.98 |
| Consensus MFE | -11.53 |
| Energy contribution | -11.89 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.873056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9067876 106 + 24543557 -----AAAUGAUGGCCAAGUGGCUCCUUGGCCUUUGGCUGGCGAUCCUGCUAGUG---GAUGUUAGUGUAGAAAGUUUAUUAAUAAUUUCAAAUUGUGUUUUU--UAAGGAUAUCA -----.......(((((((......)))))))....(((((((....))))))).---((((((....(((((((..((...............))..)))))--))..)))))). ( -26.26, z-score = -0.89, R) >droSim1.chr3L 8506300 100 + 22553184 -----AAAUGAUG-CCAAGUGGCUCUU--GCCUUUG-CUGCGAUCUCUGCUAGUG---GAUGUUAGUGUAGAAAGUUUAU-AAUAAUUUCAGUU-GUGCUAUU--AAAGGAUAAUA -----....((.(-((....)))))..--.((((((-..((..(((..(((((..---....)))))..)))........-.(((((....)))-))))...)--)))))...... ( -17.40, z-score = 1.11, R) >droSec1.super_0 1338270 107 + 21120651 -----AAAUGAUGGCCAAGUGGCUCCUUGGCCUUUGGCUGGCGAUCCUGCUAGUG---GAUGUUAGUGUAGAAAGUUUAUUAAUAAUUUCAGUUUGUGUUUUUG-AAAGGAUAAUA -----.......(((((((......))))))).((.(((((((....))))))).---))((((((((.........)))))))).((((((.........)))-)))........ ( -26.40, z-score = -0.95, R) >droYak2.chr3L 21354835 83 - 24197627 --------ACAUGGCCAAGUGGCUCCUUGGCCUUUGGCUGGCGAUCCUGCUAGUG---GAUGUUAGUGUACAAAAUUUAUUCAUAAUUUAAUCU---------------------- --------(((((((((((......))))))).((.(((((((....))))))).---)).....)))).........................---------------------- ( -23.10, z-score = -1.32, R) >droEre2.scaffold_4784 20792520 105 - 25762168 --------AUAUGGCCAAGUGGCUCCUUGGCCUUUGGCUGGCGAUCCUGCUAGUG---GACGUUAGUAUCGAAAGUUUAUUAAUAAUUUCACCUUGCUUUUUUUUGAAGAACAAUA --------....(((((((......))))))).((.(((((((....))))))).---)).(((((((.(....)..)))))))............((((.....))))....... ( -27.00, z-score = -1.15, R) >droAna3.scaffold_13337 19478337 99 + 23293914 --------AUAUGGCCAAGUGGCUCCUGAGCCUCUGGCUGGCGGUUCUGCUGGUG---GAUGUUAGUAUUUU---UGUGUUUGUGACUUUAUUU---GCAUGAAAAAAACAUAAUA --------.((((.((.((.((((....)))).)).((..(((....)))..)))---).........((((---..(((..(((....)))..---)))..))))...))))... ( -25.30, z-score = -0.76, R) >droGri2.scaffold_15110 21832361 100 - 24565398 UAUAGAGUCAAUGAUAUAUAGGGUUUUAUGCCUUGGACUGGCACUCCUUUUUGUGCUGGUCGUGAGUAUUUCAA-UUGAU--AUCAAUUUACUUAGACUGACA------------- .....((((.........((((((.....))))))(((..((((........))))..))).((((((....((-(((..--..)))))))))))))))....------------- ( -29.40, z-score = -3.03, R) >consensus _____A__AGAUGGCCAAGUGGCUCCUUGGCCUUUGGCUGGCGAUCCUGCUAGUG___GAUGUUAGUGUAGAAAGUUUAUUAAUAAUUUCACUU_GUGUUUUU__AAAGGAUAAUA ............(((((((......)))))))....(((((((....)))))))....((.((((..((((.....))))...)))).)).......................... (-11.53 = -11.89 + 0.36)

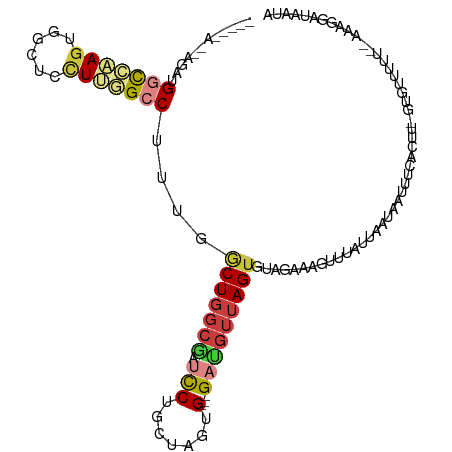
| Location | 9,067,876 – 9,067,982 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 63.70 |
| Shannon entropy | 0.70891 |
| G+C content | 0.38805 |
| Mean single sequence MFE | -16.67 |
| Consensus MFE | -8.51 |
| Energy contribution | -8.96 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.57 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.879923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9067876 106 - 24543557 UGAUAUCCUUA--AAAAACACAAUUUGAAAUUAUUAAUAAACUUUCUACACUAACAUC---CACUAGCAGGAUCGCCAGCCAAAGGCCAAGGAGCCACUUGGCCAUCAUUU----- ((((.......--..........................................(((---(.......))))...........(((((((......)))))))))))...----- ( -17.90, z-score = -1.39, R) >droSim1.chr3L 8506300 100 - 22553184 UAUUAUCCUUU--AAUAGCAC-AACUGAAAUUAUU-AUAAACUUUCUACACUAACAUC---CACUAGCAGAGAUCGCAG-CAAAGGC--AAGAGCCACUUGG-CAUCAUUU----- ......(((((--..(.((.(-....)........-........(((...(((.....---...)))...)))..))).-.))))).--..(((((....))-).))....----- ( -10.10, z-score = 0.79, R) >droSec1.super_0 1338270 107 - 21120651 UAUUAUCCUUU-CAAAAACACAAACUGAAAUUAUUAAUAAACUUUCUACACUAACAUC---CACUAGCAGGAUCGCCAGCCAAAGGCCAAGGAGCCACUUGGCCAUCAUUU----- (((((...(((-((...........)))))....)))))................(((---(.......))))...........(((((((......))))))).......----- ( -18.00, z-score = -1.90, R) >droYak2.chr3L 21354835 83 + 24197627 ----------------------AGAUUAAAUUAUGAAUAAAUUUUGUACACUAACAUC---CACUAGCAGGAUCGCCAGCCAAAGGCCAAGGAGCCACUUGGCCAUGU-------- ----------------------.........((..((.....))..)).....((((.---.....((.((....)).))....(((((((......)))))))))))-------- ( -17.20, z-score = -0.73, R) >droEre2.scaffold_4784 20792520 105 + 25762168 UAUUGUUCUUCAAAAAAAAGCAAGGUGAAAUUAUUAAUAAACUUUCGAUACUAACGUC---CACUAGCAGGAUCGCCAGCCAAAGGCCAAGGAGCCACUUGGCCAUAU-------- ..(((((.((.....)).)))))(((((......................(....)((---(.......)))))))).......(((((((......)))))))....-------- ( -22.50, z-score = -1.24, R) >droAna3.scaffold_13337 19478337 99 - 23293914 UAUUAUGUUUUUUUCAUGC---AAAUAAAGUCACAAACACA---AAAAUACUAACAUC---CACCAGCAGAACCGCCAGCCAGAGGCUCAGGAGCCACUUGGCCAUAU-------- ...((((.....(((.(((---...................---..............---.....))))))......(((((.((((....)))).).)))))))).-------- ( -14.06, z-score = 0.01, R) >droGri2.scaffold_15110 21832361 100 + 24565398 -------------UGUCAGUCUAAGUAAAUUGAU--AUCAA-UUGAAAUACUCACGACCAGCACAAAAAGGAGUGCCAGUCCAAGGCAUAAAACCCUAUAUAUCAUUGACUCUAUA -------------.((((((.......(((((..--..)))-))((.(((.....(((..((((........))))..)))..(((........))).))).))))))))...... ( -16.90, z-score = -1.02, R) >consensus UAUUAUCCUUU__AAAAACAC_AAAUGAAAUUAUUAAUAAACUUUCUACACUAACAUC___CACUAGCAGGAUCGCCAGCCAAAGGCCAAGGAGCCACUUGGCCAUCA__U_____ ....................................................................................(((((((......)))))))............ ( -8.51 = -8.96 + 0.45)
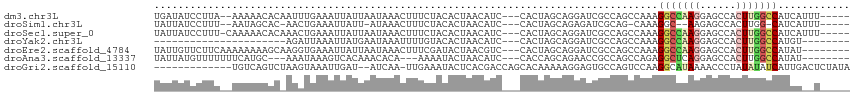
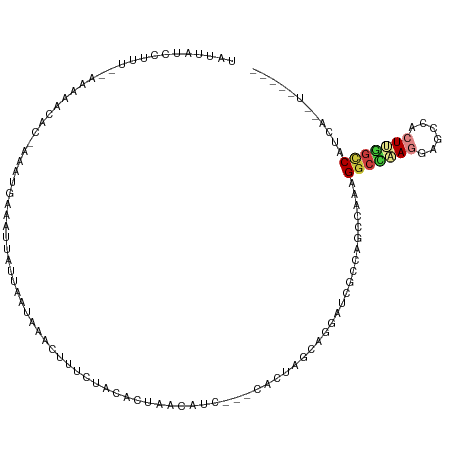
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:12:53 2011