| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,064,828 – 9,064,945 |
| Length | 117 |
| Max. P | 0.555856 |

| Location | 9,064,828 – 9,064,945 |
|---|---|
| Length | 117 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.69 |
| Shannon entropy | 0.46676 |
| G+C content | 0.42434 |
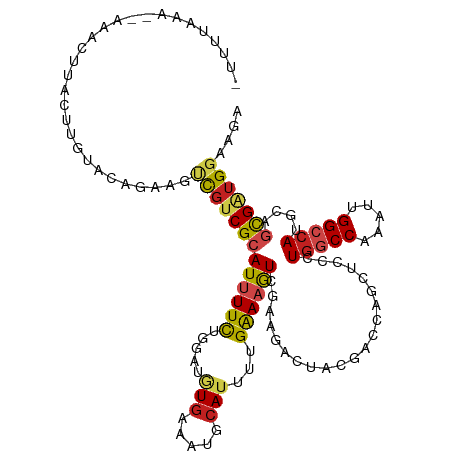
| Mean single sequence MFE | -31.71 |
| Consensus MFE | -15.83 |
| Energy contribution | -15.99 |
| Covariance contribution | 0.16 |
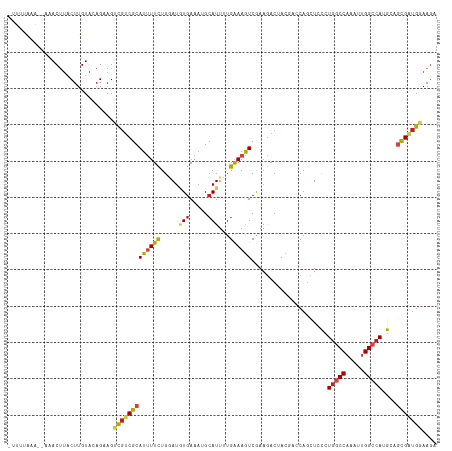
| Combinations/Pair | 1.38 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.555856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
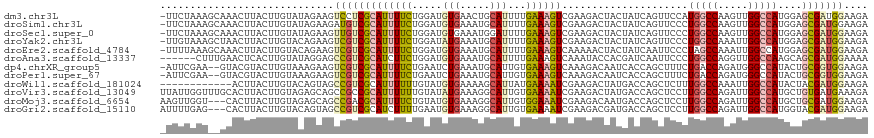
>dm3.chr3L 9064828 117 + 24543557 -UUCUAAAGCAAACUUACUUGUAUAGAAGUCCUCGCAUUUUCUGGAUGUGAACUGCAUUUUGAAAGUCGAAGACUACUAUCAGUUCCAUGGCCAAGUUGGCCAUGGAGCGAUGGAAGA -.((((..((((......)))).))))((((.(((.((((((..(((((.....)))))..))))))))).)))).(((((.(((((((((((.....)))))))))))))))).... ( -47.70, z-score = -6.07, R) >droSim1.chr3L 8503348 117 + 22553184 -UUCUAAAGCAAACUUACUUGUAUAGAAGAUGUCGCAUUUUCUGGAUGUGAAAUGCAUUUUGAAAGUCGAAGACUACUAUCAGUUCCCUGGCCAAGUUGGCCAUGGAGCGAUGGAAGA -(((((..((((......)))).)))))....(((.((((((..(((((.....)))))..)))))))))...((.(((((.(((((.(((((.....))))).)))))))))).)). ( -39.50, z-score = -3.86, R) >droSec1.super_0 1335146 117 + 21120651 -UUCUAAAGCAAACUUACUUGUAUAGAAGUUGUCGCAUUUUCUGGAUGUGAAAUGGAUUUUGAAAGUCGAAGACUACUAUCAGUUCCCUGGCCAAGUUGGCCAUGGAGCGAUGGAAGA -.((((..((((......)))).))))((((.(((.((((((..(((.(.....).)))..))))))))).)))).(((((.(((((.(((((.....))))).)))))))))).... ( -37.50, z-score = -3.14, R) >droYak2.chr3L 21351732 117 - 24197627 -UUGUAAAGCUAACUUACUUGUACAGAAGUCGUCGCAUUUUCUGGAUAUGAAAUGCAUUUUGAAAGUCGAAGACUACUAUCAGUUCCCUGGCCAAAUUGGCCAUGGAGCGAUGGAAGA -((((((((........))).))))).((((.(((.((((((.....(((.....)))...))))))))).)))).(((((.(((((.(((((.....))))).)))))))))).... ( -36.90, z-score = -3.24, R) >droEre2.scaffold_4784 20789460 117 - 25762168 -UUUUAAAGCAAACUUACUUGUACAGAAGUCGUCGCAUUUUCUGGAUGUGAAAUGCAUUUUGAAAGUCAAAAACUACUAUCAAUUCCCUAGCCAAAUUGGCCAUGGAGCGAUGGAAGA -.......((((......)))).......(((((((((((((..(((((.....)))))..)))))).................(((...(((.....)))...)))))))))).... ( -26.50, z-score = -1.06, R) >droAna3.scaffold_13337 20908584 112 + 23293914 ------CUUUGAACUCACUUGUAUAGGAGCCGUCGCAUCUUCUGGAUGUGAAAUGCAUUUUGAAAGUCAAAUACCACGAUCAAUUCCCUGGCCAGGUUGGCCAAGCAGCGAUGGAAAA ------.......(((.((.....)))))(((((((...(((..(((((.....)))))..)))........................(((((.....)))))....))))))).... ( -29.40, z-score = -0.48, R) >dp4.chrXR_group5 502163 115 + 740492 -AUUCGAA--GUACGUACUUGUAAAGAAGUCGUCGCAUUUUCUGAAUCUGAAAUGCAUUGUGAAAGUCAAAGACAAUCACCAGCUUUCUGACCAGAUGGGCCAUACUGCGGUGGAAGA -.((((..--(.(((.((((......))))))))(((((((........)))))))....)))).(((...)))..(((((.((....((.((....))..))....))))))).... ( -22.70, z-score = 1.31, R) >droPer1.super_67 91047 115 - 321716 -AUUCGAA--GUACGUACUUGUAAAGAAGUCGUCGCAUUUUCUGAAUCUGAAAUGCAUUGUGAAAGUCAAAGACAAUCACCAGCUUUCUGACCAGAUGGGCCAUACUGCGGUGGAAGA -.((((..--(.(((.((((......))))))))(((((((........)))))))....)))).(((...)))..(((((.((....((.((....))..))....))))))).... ( -22.70, z-score = 1.31, R) >droWil1.scaffold_181024 1066983 106 + 1751709 ------------ACUUACUUGUACAGUAGCCGUCGCAUUUUUUGUAUGUGAAAAGCAUUAUGAAAAUCGAAGACUAUGACCAGCUCUUUGGCCAAAUUGGCCAUACUACGAUGGAAGA ------------.....(((.....((((.(.(((.((((((...((((.....))))...))))))))).).))))..(((......(((((.....)))))........)))))). ( -23.64, z-score = -0.92, R) >droVir3.scaffold_13049 23586660 118 - 25233164 UUAUUGGUUUGCACUUACUUGUAGAGCAGCCGCCGCAUUUUUUGUAUAUGAAAGGCAUUGUGAAAAUCGAAGACUAUGACCAGCUCCUUGGCCAGAUUGGCCAUGCUGUGAUGAAAGA ...(((((((.(((....(((.....)))..(((.(((.........)))...)))...))).)))))))....(((.((.(((....(((((.....))))).))))).)))..... ( -27.60, z-score = 0.25, R) >droMoj3.scaffold_6654 2172589 115 + 2564135 AAGUUGGU---CACUUACUUGUAGAGCAGCCGACGCAUUUUCUGUAUGUGAAAGGCAUUGUGGAAAUCGAAGACAAUGACCAGCUCCUUGGCCAGAUUGGCCAUGCUGCGAUGGAAGA ..((((((---..(((.......)))..))))))...((((((((.(((....((((((((...........)))))).))(((....(((((.....))))).)))))))))))))) ( -33.60, z-score = -0.59, R) >droGri2.scaffold_15110 21829227 115 - 24565398 AUUUUGAG---CACUUACUUGUACAGUAGCCGUCGCAUCUUUUGAAUGUGAAAGGCAUUGUGAAAAUCGAAGACGAUGACCAGCUCCUUGGCCAGAUUGGCCAUGGUACGAUGGAAGA .((((((.---(((.((((.....))))(((.(((((((....).))))))..)))...)))....)))))).......(((...((.(((((.....))))).)).....))).... ( -32.80, z-score = -1.42, R) >consensus _UUUUAAA__AAACUUACUUGUACAGAAGUCGUCGCAUUUUCUGGAUGUGAAAUGCAUUUUGAAAGUCGAAGACUACGACCAGCUCCCUGGCCAAAUUGGCCAUGCAGCGAUGGAAGA .............................(((((((((((((.....(((.....)))...)))))).....................(((((.....)))))....))))))).... (-15.83 = -15.99 + 0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:12:51 2011