
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,057,785 – 9,057,884 |
| Length | 99 |
| Max. P | 0.981615 |

| Location | 9,057,785 – 9,057,880 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 87.49 |
| Shannon entropy | 0.20777 |
| G+C content | 0.42041 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -22.30 |
| Energy contribution | -22.10 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.981615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
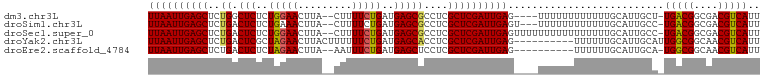
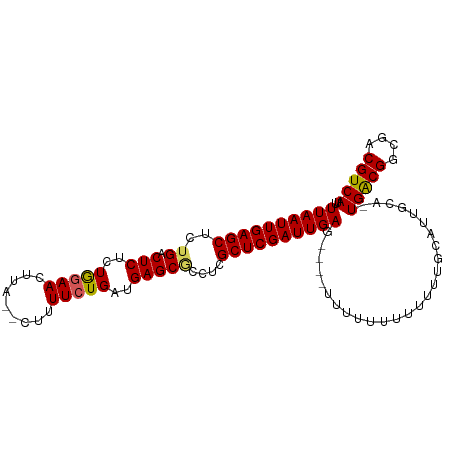
>dm3.chr3L 9057785 95 - 24543557 UUAAUUGAGCUCUGGCUCUCUGGAACUUA--CUUUUCUGAUGAGCGCCUCGCUCGAUUGAG----UUUUUUUUUUUUGCAUUGCU-UGACGGCGACGUCAUU ((((((((((...(((.(((.((((....--...))))...))).)))..)))))))))).----....................-(((((....))))).. ( -29.50, z-score = -3.30, R) >droSim1.chr3L 8496317 96 - 22553184 UUAAUUGAGCUCUGACUCUCUGAAACUUA--CUUUUCUGAUGAGCGCCUCGCUCGAUUGAGU---UUUUUUUUUUUUGCAUUGCC-UGACGGCGACGUCAUU ((((((((((..((.(((((.((((....--..)))).)).)))))....))))))))))..---....................-(((((....))))).. ( -23.90, z-score = -2.18, R) >droSec1.super_0 1328173 99 - 21120651 UUAAUUGAGCUCUGACUCUCUGGAACUUA--CUUUUCUGAUGAGCGCCUCGCUCGAUUGAGUUUUUUUUUUUUUUUUGCAUUGCC-UGACGGCGACGUCAUU ((((((((((..((.(((((.((((....--..)))).)).)))))....)))))))))).........................-(((((....))))).. ( -23.40, z-score = -1.75, R) >droYak2.chr3L 21344788 92 + 24197627 UUAAUUGAGCUCUGACUCGCUAGAACUUACUUUUUUCUGAUGAGCACCUCGCUCGAUUGAG----------UUUUUUGCAUUGCAUUGGCGGCAACGUCAUU ((((((((((..((.((((.(((((.........))))).))))))....)))))))))).----------.....(((...))).(((((....))))).. ( -25.80, z-score = -2.27, R) >droEre2.scaffold_4784 20782527 89 + 25762168 UUAAUUGAGCUCUGACUCUCUAGAACUUA--AAUUUCUGAUGAGCUCCUCGCUCGAUUGAG----------UUUUUUGCAUUGCA-UGGCGGCAACGUCAUU ((((((((((...(((((..(((((....--...)))))..))).))...)))))))))).----------.............(-(((((....)))))). ( -27.00, z-score = -2.81, R) >consensus UUAAUUGAGCUCUGACUCUCUGGAACUUA__CUUUUCUGAUGAGCGCCUCGCUCGAUUGAG____UUUUUUUUUUUUGCAUUGCA_UGACGGCGACGUCAUU ((((((((((..((.(((..(((((.........)))))..)))))....))))))))))..........................(((((....))))).. (-22.30 = -22.10 + -0.20)
| Location | 9,057,788 – 9,057,884 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 87.61 |
| Shannon entropy | 0.20575 |
| G+C content | 0.44754 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -22.08 |
| Energy contribution | -21.88 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.937431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
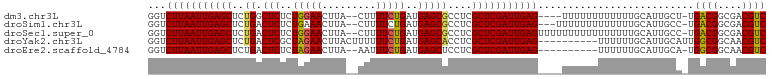
>dm3.chr3L 9057788 96 - 24543557 GGUCUUAAUUGAGCUCUGGCUCUCUGGAACUUA--CUUUUCUGAUGAGCGCCUCGCUCGAUUGAG----UUUUUUUUUUUUGCAUUGCU-UGACGGCGACGUC ...(((((((((((...(((.(((.((((....--...))))...))).)))..)))))))))))----....................-.((((....)))) ( -29.60, z-score = -2.87, R) >droSim1.chr3L 8496320 97 - 22553184 GGUCUUAAUUGAGCUCUGACUCUCUGAAACUUA--CUUUUCUGAUGAGCGCCUCGCUCGAUUGAG---UUUUUUUUUUUUUGCAUUGCC-UGACGGCGACGUC ...(((((((((((..((.(((((.((((....--..)))).)).)))))....)))))))))))---.............((.(((((-....))))).)). ( -25.20, z-score = -2.07, R) >droSec1.super_0 1328176 100 - 21120651 GGUCUUAAUUGAGCUCUGACUCUCUGGAACUUA--CUUUUCUGAUGAGCGCCUCGCUCGAUUGAGUUUUUUUUUUUUUUUUGCAUUGCC-UGACGGCGACGUC ...(((((((((((..((.(((((.((((....--..)))).)).)))))....)))))))))))................((.(((((-....))))).)). ( -24.70, z-score = -1.63, R) >droYak2.chr3L 21344791 93 + 24197627 GGUCUUAAUUGAGCUCUGACUCGCUAGAACUUACUUUUUUCUGAUGAGCACCUCGCUCGAUUGAG----------UUUUUUGCAUUGCAUUGGCGGCAACGUC ...(((((((((((..((.((((.(((((.........))))).))))))....)))))))))))----------.....(((...)))..((((....)))) ( -25.90, z-score = -1.77, R) >droEre2.scaffold_4784 20782530 90 + 25762168 GGUCUUAAUUGAGCUCUGACUCUCUAGAACUUA--AAUUUCUGAUGAGCUCCUCGCUCGAUUGAG----------UUUUUUGCAUUGCA-UGGCGGCAACGUC ...(((((((((((...(((((..(((((....--...)))))..))).))...)))))))))))----------.....(((...)))-.((((....)))) ( -26.50, z-score = -2.18, R) >consensus GGUCUUAAUUGAGCUCUGACUCUCUGGAACUUA__CUUUUCUGAUGAGCGCCUCGCUCGAUUGAG____UUUUUUUUUUUUGCAUUGCA_UGACGGCGACGUC ...(((((((((((..((.(((..(((((.........)))))..)))))....)))))))))))..........................((((....)))) (-22.08 = -21.88 + -0.20)
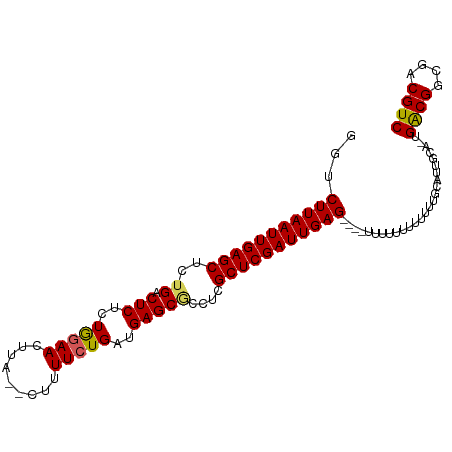
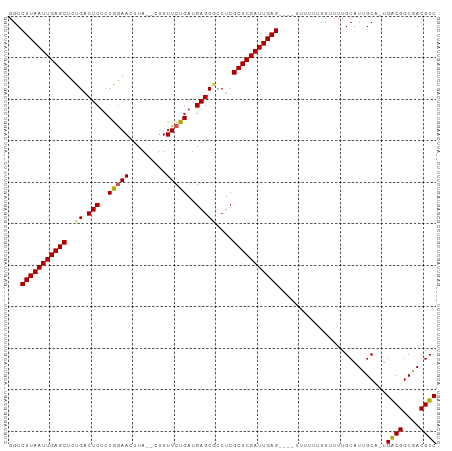
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:12:50 2011