| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,031,178 – 9,031,272 |
| Length | 94 |
| Max. P | 0.989531 |

| Location | 9,031,178 – 9,031,272 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 84.75 |
| Shannon entropy | 0.31877 |
| G+C content | 0.49298 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -24.94 |
| Energy contribution | -24.84 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.989531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
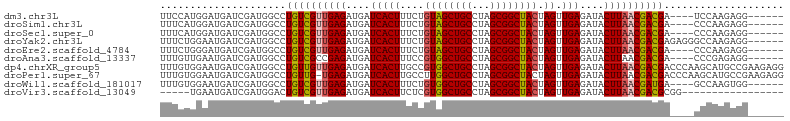
>dm3.chr3L 9031178 94 - 24543557 UUCCAUGGAUGAUCGAUGGCCUGUCGUUGAGAUGAUCACUUUCUGUAGCUGCCUAGCGGCUACUAGUUGAGAUACUUAACGACGA----UCCAAGAGG------ ..((((.(.....).))))..((((((((((....(((((....((((((((...)))))))).)).)))....)))))))))).----.........------ ( -31.50, z-score = -2.71, R) >droSim1.chr3L 8469386 94 - 22553184 UUUCAUGGAUGAUCGAUGGCCUGUCGUUGAGAUGAUCACUUUCUGUAGCUGCCUAGCGGCUACUAGUUGAGAUACUUAACGACGA----CCCAAGAGG------ ............((..(((..((((((((((....(((((....((((((((...)))))))).)).)))....)))))))))).----.))).))..------ ( -31.50, z-score = -2.99, R) >droSec1.super_0 1301672 94 - 21120651 UUUCAUGGAUGAUCGAUGGCCUGUCGUUGAGAUGAUCACUUUCUGUAGCUGCCUAGCGGCUACUAGUUGAGAUACUUAACGACGA----CCCAAGAGG------ ............((..(((..((((((((((....(((((....((((((((...)))))))).)).)))....)))))))))).----.))).))..------ ( -31.50, z-score = -2.99, R) >droYak2.chr3L 21317580 98 + 24197627 UUUCUGGAAUGAUCGAUGGCCUGUCGUUGAGAUGAUCACUUUCUGUAGCUGCCUAGCGGCUACUAGUUGAGAUACUUAACGACGAGAGGGCCAAGAGG------ ............((..(((((((((((((((....(((((....((((((((...)))))))).)).)))....)))))))))....)))))).))..------ ( -36.50, z-score = -3.34, R) >droEre2.scaffold_4784 20755768 94 + 25762168 UUUCUGGGAUGAUCGAUGGCCUGUCGUUGAGAUGAUCACUUUCUGUAGCUGCCUAGCGGCUACUAGUUGAGAUACUUAACGACGA----CCCAAGAGG------ .(((((((......(.....)((((((((((....(((((....((((((((...)))))))).)).)))....)))))))))).----))).)))).------ ( -31.90, z-score = -2.78, R) >droAna3.scaffold_13337 15672888 94 + 23293914 UUUGUUGAAUGAUCGAUGGCCUGUCGCCGAGAUGAUCACUUUCCGUGGCUGCCUAGCGGCUACUAGUUGAGAUACUUAACGACGA----CCCGAGAGG------ .........((((((.((((.....))))...))))))(((((.((((((((...))))))))..((((((...)))))).....----...))))).------ ( -28.80, z-score = -1.94, R) >dp4.chrXR_group5 467292 104 - 740492 UUUGUGGAAUGAUCGAUGGCCUGUUGUUGAGAUGAUCACUUGCCGUGGCUGCCUAGCGGCUACUAGUUGAGAUACUUAACGACGACCCAAGCAUGCCGAAGAGG ....(((.(((.....(((..((((((((((....(((((....((((((((...)))))))).)).)))....))))))))))..)))..))).)))...... ( -31.20, z-score = -1.80, R) >droPer1.super_67 56156 103 + 321716 UUUGUGGAAUGAUCGAUGGCCUGUUG-UGAGAUGAUCACUUGCCUUGGCUGCCUAGCGGCUACUAGUUGAGAUACUUAACGACGACCCAAGCAUGCCGAAGAGG ............((..((((.(((((-(((.....))))......(((((((...)))))))...((((((...)))))).........)))).))))..)).. ( -24.70, z-score = -0.04, R) >droWil1.scaffold_181017 638720 94 - 939937 UUUGUGGAAUGAUCGAUGGCCUGUCGUUGAGAUGAUCACUUUCUGUGGCUGCCUAGCGGCUACUAGUUGAGAUACUUAACGAUGA----GCCAAGUGG------ ................((((..(((((((((....(((((....((((((((...)))))))).)).)))....)))))))))..----)))).....------ ( -33.10, z-score = -3.25, R) >droVir3.scaffold_13049 8770984 82 + 25233164 -----UGAAUGAUCGAUGGACUGUCGUUGAGAUGAUCACUUCUCGUGGCUGCCUAGCGGCUACUAGUUGAGAUACUUAACGACGCGG----------------- -----.................(((((((((...(((.(..((.((((((((...)))))))).))..).))).)))))))))....----------------- ( -27.30, z-score = -2.52, R) >consensus UUUCUGGAAUGAUCGAUGGCCUGUCGUUGAGAUGAUCACUUUCUGUAGCUGCCUAGCGGCUACUAGUUGAGAUACUUAACGACGA____CCCAAGAGG______ .....................((((((((((....(((((....((((((((...)))))))).)).)))....)))))))))).................... (-24.94 = -24.84 + -0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:12:46 2011