
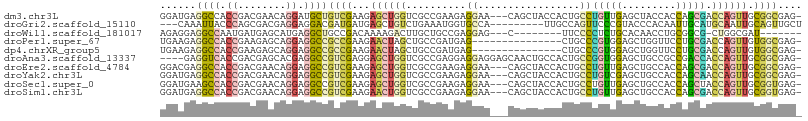
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,987,586 – 8,987,689 |
| Length | 103 |
| Max. P | 0.594628 |

| Location | 8,987,586 – 8,987,689 |
|---|---|
| Length | 103 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 66.88 |
| Shannon entropy | 0.70836 |
| G+C content | 0.62241 |
| Mean single sequence MFE | -34.71 |
| Consensus MFE | -14.15 |
| Energy contribution | -14.58 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.65 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.594628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
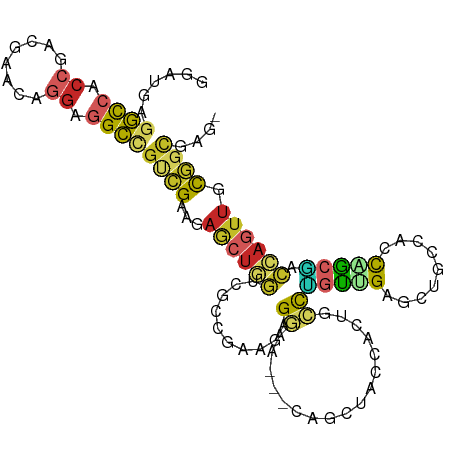
>dm3.chr3L 8987586 103 + 24543557 GGAUGAGGCCACCGACGAACAGGAUGCUGUCGAAGAGCUGGUCGCCGAAGAGGAA---CAGCUACCACUGCCUGUUGAGCUACCACCAGCGACCAGUUGCGGCGAG- .......(((.....(((.(((....))))))...((((((((((.(....((..---.((((.(.((.....)).))))).))..).))))))))))..)))...- ( -32.40, z-score = -0.37, R) >droGri2.scaffold_15110 12183724 95 + 24565398 ---CAAAUUACCCAGCGACGAGGAGGACGAUGAUGAGCUGUCUGAAAUGGUGCCA---------UUGCCAGUUCCCGUACCCACAAUUGCAUGCAAUUGCAGUUGCU ---..........((((((..((.(.(((.....((((((.....((((....))---------))..)))))).))).))).((((((....))))))..)))))) ( -24.30, z-score = -0.29, R) >droWil1.scaffold_181017 576358 88 + 939937 AGAGGAGGCCAAUGAUGAGCAUGAGGCUGCCGACAAAAGACUUGCUGCCGAGGAG---C--------UUCCCCUCUGCACAACCUGCGGCG-CUGGCGAU------- .......((((......(((.....)))((((.....((..(((.(((.((((..---.--------....)))).)))))).)).)))).-.))))...------- ( -25.50, z-score = 1.00, R) >droPer1.super_67 13644 91 - 321716 UGAAGAGGCCACCGAAGAGCAGGAGGCCGCCGAAGAACUAGCUGCCGAUGAG---------------CUGCCCGUGGAGCUGGUUCCUGCGACCAGUUGUGGCGAG- ......((((.((........)).))))(((...(((((((((.((.(((..---------------.....))))))))))))))..((((....)))))))...- ( -34.50, z-score = -1.56, R) >dp4.chrXR_group5 425177 91 + 740492 UGAAGAGGCCACCGAAGAGCAGGAGGCCGCCGAAGAACUAGCUGCCGAUGAG---------------CUGCCCGUGGAGCUGGUUCCUGCGACCAGUUGUGGCGAG- ......((((.((........)).))))(((...(((((((((.((.(((..---------------.....))))))))))))))..((((....)))))))...- ( -34.50, z-score = -1.56, R) >droAna3.scaffold_13337 15630196 102 - 23293914 ----GAGGUCACCGACGAGCACGAGGCCGUCGAGGAGCUGGUCGCCGAGGAGGAGGAGCAACUGCCACUGCCGGUGGAGCUGCCGCCGACCACCAGUUGCGGCGAG- ----.((.((..(((((.((.....)))))))..)).))..((((((.((.((((..((....))..))..((((((.....)))))).)).)).....)))))).- ( -42.90, z-score = -0.29, R) >droEre2.scaffold_4784 20709199 103 - 25762168 GGACGAGGCCACCGACGAACAGGAGGCCGUCGAAGAGCUGGUCGCCGAAGAGGAA---CAGCUACCACUGCCUGUUGAGCUGCCACCAGCGACCAGUUGCGGCGAG- ......((((.((........)).))))((((...((((((((((.(....((..---(((((.(.((.....)).))))))))..).)))))))))).))))...- ( -41.40, z-score = -1.94, R) >droYak2.chr3L 21271898 103 - 24197627 GGAUGAGGCCACCGACGAACAGGAGGCCGUCGAAGAGCUGGUCGCCGAAGAGGAA---CAGCUACCACUGCCUGUCGAGCUGCCACCAGCAACCAGUUGCGGCGAG- ......((((.((........)).))))((((...(((((((...(((..(((..---(((......)))))).))).((((....)))).))))))).))))...- ( -36.60, z-score = -0.66, R) >droSec1.super_0 1258326 103 + 21120651 GGAUGAAGCCACCGACGAACAGGAGGCCGUCGAAGAGCUGGUCGCCGAAGAGGAA---CAGCUACCACUGCCUGUUGAGCUGCCACCAGCUACCAGUUGCGGUGAG- .((((..(((.((........)).)))))))....(((((....((.....))..---)))))..(((((((((...(((((....)))))..)))..))))))..- ( -36.60, z-score = -1.40, R) >droSim1.chr3L 8425564 103 + 22553184 GGAUGAGGCCACCGACGAACAGGAGGCCGUCGAAGAACUGGUCGCCGAAGAGGAA---CAGCUACCACUGCCUGUUGAGCUGCCACCAGCGACCAGUUGCGGUGAG- ......((((.((........)).)))).(((..(((((((((((.(....((..---(((((.(.((.....)).))))))))..).)))))))))).)..))).- ( -38.40, z-score = -1.79, R) >consensus GGAUGAGGCCACCGACGAACAGGAGGCCGUCGAAGAGCUGGUCGCCGAAGAGGAA___CAGCUACCACUGCCUGUUGAGCUGCCACCAGCGACCAGUUGCGGCGAG_ ......((((.((........)).))))((((...((((((.(((.(.......................................).))).)))))).)))).... (-14.15 = -14.58 + 0.43)

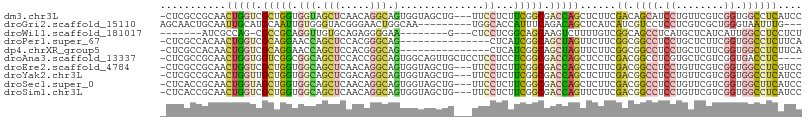
| Location | 8,987,586 – 8,987,689 |
|---|---|
| Length | 103 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 66.88 |
| Shannon entropy | 0.70836 |
| G+C content | 0.62241 |
| Mean single sequence MFE | -36.80 |
| Consensus MFE | -13.41 |
| Energy contribution | -13.48 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.78 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.527368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8987586 103 - 24543557 -CUCGCCGCAACUGGUCGCUGGUGGUAGCUCAACAGGCAGUGGUAGCUG---UUCCUCUUCGGCGACCAGCUCUUCGACAGCAUCCUGUUCGUCGGUGGCCUCAUCC -...(((((..(((((((((((.((((((((.((.....)).).)))))---..))...))))))))))).....(((((((.....))).)))))))))....... ( -40.80, z-score = -2.22, R) >droGri2.scaffold_15110 12183724 95 - 24565398 AGCAACUGCAAUUGCAUGCAAUUGUGGGUACGGGAACUGGCAA---------UGGCACCAUUUCAGACAGCUCAUCAUCGUCCUCCUCGUCGCUGGGUAAUUUG--- (((..(..((((((....))))))..)(.((((((.((((.((---------((....))))))))...((........))..))).)))))))..........--- ( -23.50, z-score = 0.57, R) >droWil1.scaffold_181017 576358 88 - 939937 -------AUCGCCAG-CGCCGCAGGUUGUGCAGAGGGGAA--------G---CUCCUCGGCAGCAAGUCUUUUGUCGGCAGCCUCAUGCUCAUCAUUGGCCUCCUCU -------...(((((-.((((((((((((((.((((((..--------.---)))))).))).)))....)))).))))(((.....))).....)))))....... ( -28.30, z-score = 0.70, R) >droPer1.super_67 13644 91 + 321716 -CUCGCCACAACUGGUCGCAGGAACCAGCUCCACGGGCAG---------------CUCAUCGGCAGCUAGUUCUUCGGCGGCCUCCUGCUCUUCGGUGGCCUCUUCA -...(((((....((..((((((...((((((...)).))---------------))....(((.(((........))).))))))))).))...)))))....... ( -31.70, z-score = -0.83, R) >dp4.chrXR_group5 425177 91 - 740492 -CUCGCCACAACUGGUCGCAGGAACCAGCUCCACGGGCAG---------------CUCAUCGGCAGCUAGUUCUUCGGCGGCCUCCUGCUCUUCGGUGGCCUCUUCA -...(((((....((..((((((...((((((...)).))---------------))....(((.(((........))).))))))))).))...)))))....... ( -31.70, z-score = -0.83, R) >droAna3.scaffold_13337 15630196 102 + 23293914 -CUCGCCGCAACUGGUGGUCGGCGGCAGCUCCACCGGCAGUGGCAGUUGCUCCUCCUCCUCGGCGACCAGCUCCUCGACGGCCUCGUGCUCGUCGGUGACCUC---- -.((((((.....((.((..((.(((((((((((.....)))).))))))))).)).)).))))))..((.((.((((((((.....)).)))))).)).)).---- ( -42.50, z-score = -0.50, R) >droEre2.scaffold_4784 20709199 103 + 25762168 -CUCGCCGCAACUGGUCGCUGGUGGCAGCUCAACAGGCAGUGGUAGCUG---UUCCUCUUCGGCGACCAGCUCUUCGACGGCCUCCUGUUCGUCGGUGGCCUCGUCC -...(((((..(((((((((((.((((((((.((.....)).).)))))---..))...))))))))))).....(((((..(....)..))))))))))....... ( -45.10, z-score = -2.38, R) >droYak2.chr3L 21271898 103 + 24197627 -CUCGCCGCAACUGGUUGCUGGUGGCAGCUCGACAGGCAGUGGUAGCUG---UUCCUCUUCGGCGACCAGCUCUUCGACGGCCUCCUGUUCGUCGGUGGCCUCAUCC -...(((((..(((((((((((.((((((((.((.....)).).)))))---..))...))))))))))).....(((((..(....)..))))))))))....... ( -45.10, z-score = -2.22, R) >droSec1.super_0 1258326 103 - 21120651 -CUCACCGCAACUGGUAGCUGGUGGCAGCUCAACAGGCAGUGGUAGCUG---UUCCUCUUCGGCGACCAGCUCUUCGACGGCCUCCUGUUCGUCGGUGGCUUCAUCC -....((((....((.(((((((.((.........((((((....))))---))........)).))))))))).(((((..(....)..)))))))))........ ( -35.73, z-score = -0.40, R) >droSim1.chr3L 8425564 103 - 22553184 -CUCACCGCAACUGGUCGCUGGUGGCAGCUCAACAGGCAGUGGUAGCUG---UUCCUCUUCGGCGACCAGUUCUUCGACGGCCUCCUGUUCGUCGGUGGCCUCAUCC -....(((((((((((((((((.((((((((.((.....)).).)))))---..))...)))))))))))))...(((((..(....)..)))))))))........ ( -43.60, z-score = -2.90, R) >consensus _CUCGCCGCAACUGGUCGCUGGUGGCAGCUCAACAGGCAGUGGUAGCUG___UUCCUCUUCGGCGACCAGCUCUUCGACGGCCUCCUGUUCGUCGGUGGCCUCAUCC ...........(((((.(((((.....................................))))).)))))......((.((((.((........)).)))))).... (-13.41 = -13.48 + 0.07)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:12:45 2011