
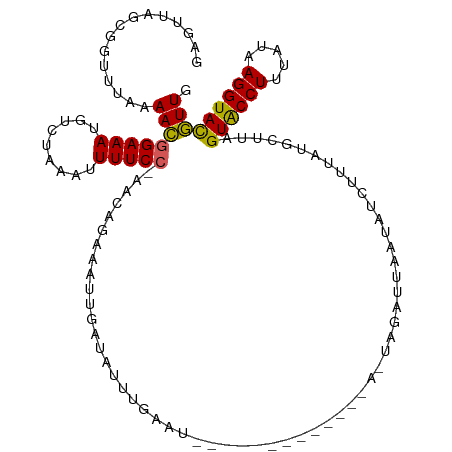
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,954,277 – 8,954,394 |
| Length | 117 |
| Max. P | 0.716726 |

| Location | 8,954,277 – 8,954,394 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 67.26 |
| Shannon entropy | 0.55918 |
| G+C content | 0.31425 |
| Mean single sequence MFE | -20.70 |
| Consensus MFE | -9.54 |
| Energy contribution | -9.66 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.716726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8954277 117 + 24543557 GAGUUAGCGGUUUAAAACGGAAAUGUCUAAAUUUUCCGAACAGAAAUUGAUAUUUGAAUUAGAUCAGUGACAAGUAGUUGAGUAUUUUUAUGCUUAGUGCCUUUAUAAGGUACGUUG ..((((..(((((((..((((((.........))))))..((((........))))..)))))))..)))).......(((((((....)))))))((((((.....)))))).... ( -26.60, z-score = -2.01, R) >droEre2.scaffold_4784 20675016 96 - 25762168 GAGUUAGCGGCUUAAAAAGGAAAUGUCGAAAUUUUCCUAACAGAUGUUCAUAUUUGGAU---------------------UAGACUACAGGUACUAGUACCUUUUAAAGGUACUUUG ....(((..((((....((((((.........))))))..((((((....))))))...---------------------........)))).)))((((((.....)))))).... ( -17.20, z-score = 0.02, R) >droYak2.chr3L 21238459 88 - 24197627 ---------------AAAAGAAAUGCCAAAAUUUUCCUAAAAGAAGUCGACUACAGGC--------------AAUAGUUUAAUAUCUUAAGGCUUAGUACCUUUUCAAGGUACUUUG ---------------.........(((......(((......)))(((.......)))--------------..................)))..(((((((.....)))))))... ( -14.50, z-score = -0.57, R) >droSec1.super_0 1224960 99 + 21120651 GAGUUAGCGGUUUAAAACGGAAAAGUCUAAAUUUUCC-AACAGAAAUUGAUAUUUGA-----------------UAGAUAAAUAUCUUUAUGCUUAGUACCUUUAUAAGGUAUGUUG .....((((.........((((((.......))))))-..........((((((((.-----------------....))))))))....))))..((((((.....)))))).... ( -17.90, z-score = -1.16, R) >droSim1.chr3L 8391568 116 + 22553184 GAGUUAGCGGUUUAAAACGGAAAUGUCUAAAUUUUCC-AACAGAAAUUGAUAUUUGAAUUAGACCACUGACAAAUAGAUUAAUAUCUUUAUGCUUAGUACCUUUAUAAGGUACGUUG ..(((((.(((((((...(((((.........)))))-..((((........))))..))))))).)))))....((((....)))).........((((((.....)))))).... ( -27.30, z-score = -3.22, R) >consensus GAGUUAGCGGUUUAAAACGGAAAUGUCUAAAUUUUCC_AACAGAAAUUGAUAUUUGAAU_____________A_UAGAUUAAUAUCUUUAUGCUUAGUACCUUUAUAAGGUACGUUG .................((((((.........))))))..........................................................((((((.....)))))).... ( -9.54 = -9.66 + 0.12)


| Location | 8,954,277 – 8,954,394 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 67.26 |
| Shannon entropy | 0.55918 |
| G+C content | 0.31425 |
| Mean single sequence MFE | -15.74 |
| Consensus MFE | -7.98 |
| Energy contribution | -8.22 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.662707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8954277 117 - 24543557 CAACGUACCUUAUAAAGGCACUAAGCAUAAAAAUACUCAACUACUUGUCACUGAUCUAAUUCAAAUAUCAAUUUCUGUUCGGAAAAUUUAGACAUUUCCGUUUUAAACCGCUAACUC ....((.(((.....))).))..(((...................((((..(((......)))........(((((....))))).....)))).....((.....)).)))..... ( -11.50, z-score = -0.08, R) >droEre2.scaffold_4784 20675016 96 + 25762168 CAAAGUACCUUUAAAAGGUACUAGUACCUGUAGUCUA---------------------AUCCAAAUAUGAACAUCUGUUAGGAAAAUUUCGACAUUUCCUUUUUAAGCCGCUAACUC ...(((((((.....))))))).......(..(..((---------------------(..........(((....)))((((((.........))))))..)))..)..)...... ( -13.80, z-score = -0.29, R) >droYak2.chr3L 21238459 88 + 24197627 CAAAGUACCUUGAAAAGGUACUAAGCCUUAAGAUAUUAAACUAUU--------------GCCUGUAGUCGACUUCUUUUAGGAAAAUUUUGGCAUUUCUUUU--------------- ...(((((((.....)))))))..(((..((........(((((.--------------....)))))....((((....))))..))..))).........--------------- ( -16.50, z-score = -0.78, R) >droSec1.super_0 1224960 99 - 21120651 CAACAUACCUUAUAAAGGUACUAAGCAUAAAGAUAUUUAUCUA-----------------UCAAAUAUCAAUUUCUGUU-GGAAAAUUUAGACUUUUCCGUUUUAAACCGCUAACUC .....(((((.....)))))...(((.....(((((((.....-----------------..)))))))..........-((((((.......))))))..........)))..... ( -15.80, z-score = -2.30, R) >droSim1.chr3L 8391568 116 - 22553184 CAACGUACCUUAUAAAGGUACUAAGCAUAAAGAUAUUAAUCUAUUUGUCAGUGGUCUAAUUCAAAUAUCAAUUUCUGUU-GGAAAAUUUAGACAUUUCCGUUUUAAACCGCUAACUC ....((((((.....))))))...(((...((((....))))...))).((((((.(((..(.((((........))))-(((((.........))))))..))).))))))..... ( -21.10, z-score = -2.06, R) >consensus CAACGUACCUUAUAAAGGUACUAAGCAUAAAGAUAUUAAACUA_U_____________AUUCAAAUAUCAAUUUCUGUU_GGAAAAUUUAGACAUUUCCGUUUUAAACCGCUAACUC ....((((((.....))))))...........................................................(((((.........))))).................. ( -7.98 = -8.22 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:12:42 2011