
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,947,753 – 8,947,817 |
| Length | 64 |
| Max. P | 0.867792 |

| Location | 8,947,753 – 8,947,817 |
|---|---|
| Length | 64 |
| Sequences | 7 |
| Columns | 66 |
| Reading direction | forward |
| Mean pairwise identity | 58.83 |
| Shannon entropy | 0.84944 |
| G+C content | 0.39142 |
| Mean single sequence MFE | -13.90 |
| Consensus MFE | -4.47 |
| Energy contribution | -4.27 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.867792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
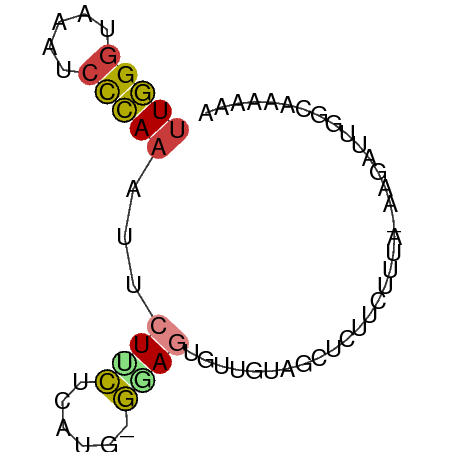
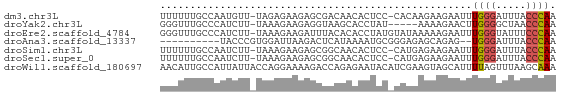
>dm3.chr3L 8947753 64 + 24543557 UUGGGUAAAUCCCAAAUUCUUCUUGUG-GGAGUGUUGUCGCUCUUCUCUA-AACAUUGGCAAAAAA (((((.....)))))...........(-((((((....))))))).....-............... ( -12.50, z-score = -1.24, R) >droYak2.chr3L 21231573 60 - 24197627 UUGGGUUAGCCCCAAGUUCUUUU-----AUAGGUGCUUACCUCUUCUUUA-AAGAUGGGCAAACCC ..(((((.((((.....(((((.-----..((((....)))).......)-)))).)))).))))) ( -17.90, z-score = -1.72, R) >droEre2.scaffold_4784 20668249 65 - 25762168 UUGGGAAAUACCCAAAUUCUUUUUAUACAUAGGUGUGUAAAUCUUCUUUA-AAGAUGGGCAAACCC (((((.....)))))................(((.(((..(((((.....-)))))..))).))). ( -15.20, z-score = -1.66, R) >droAna3.scaffold_13337 15591038 54 - 23293914 UUGGGUAAAUCCCA--CUCUGCUCUCCCGCAUUUUAUGAGUCUUAAUCCACGGGUA---------- .((((.....))))--.........((((....(((.......)))....))))..---------- ( -9.60, z-score = -0.14, R) >droSim1.chr3L 8384598 64 + 22553184 UUGGGUAAAUCCCAAAUUCUUCUCAUG-GGAGUGUUGCCGCUCUUCUUUA-AAGAUUGGCAAAAAA ...(((((.(((((...........))-)))...)))))(((..(((...-.)))..)))...... ( -17.60, z-score = -2.35, R) >droSec1.super_0 1218007 64 + 21120651 UUGGGUAAAUCCCAAAUUCUUCUCAUG-GGAGUGUUGCCGCUCUUCUUUA-AAGAUUGGCAAAAAA ...(((((.(((((...........))-)))...)))))(((..(((...-.)))..)))...... ( -17.60, z-score = -2.35, R) >droWil1.scaffold_180697 3837853 66 + 4168966 UUUGCUUAAACUAAAAUGCUACUUCGAUGUAUUCUCUGGUCUUUUCCUGGUAAUAAUGGCAAUGUU .(((((...((((.(((((.........)))))....((......))))))......))))).... ( -6.90, z-score = -0.02, R) >consensus UUGGGUAAAUCCCAAAUUCUUCUCAUG_GGAGUGUUGUAGCUCUUCUUUA_AAGAUUGGCAAAAAA (((((.....)))))...((((......)))).................................. ( -4.47 = -4.27 + -0.20)

| Location | 8,947,753 – 8,947,817 |
|---|---|
| Length | 64 |
| Sequences | 7 |
| Columns | 66 |
| Reading direction | reverse |
| Mean pairwise identity | 58.83 |
| Shannon entropy | 0.84944 |
| G+C content | 0.39142 |
| Mean single sequence MFE | -13.18 |
| Consensus MFE | -3.60 |
| Energy contribution | -3.64 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.709087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
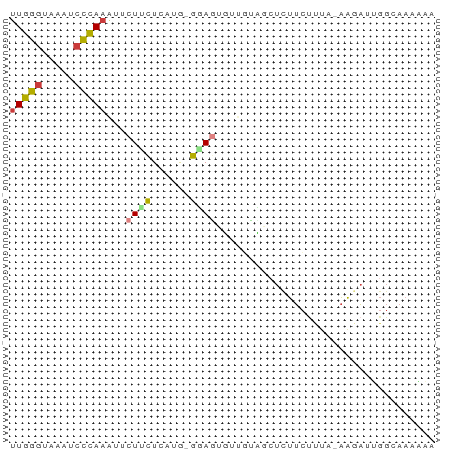
>dm3.chr3L 8947753 64 - 24543557 UUUUUUGCCAAUGUU-UAGAGAAGAGCGACAACACUCC-CACAAGAAGAAUUUGGGAUUUACCCAA (((((((.....(((-(......))))((......)).-..)))))))...(((((.....))))) ( -7.30, z-score = 0.22, R) >droYak2.chr3L 21231573 60 + 24197627 GGGUUUGCCCAUCUU-UAAAGAAGAGGUAAGCACCUAU-----AAAAGAACUUGGGGCUAACCCAA (((((.((((.....-........((((....))))..-----...........)))).))))).. ( -20.57, z-score = -2.54, R) >droEre2.scaffold_4784 20668249 65 + 25762168 GGGUUUGCCCAUCUU-UAAAGAAGAUUUACACACCUAUGUAUAAAAAGAAUUUGGGUAUUUCCCAA (((..((((((((((-(...)))))((((.(((....))).)))).......))))))...))).. ( -16.30, z-score = -2.23, R) >droAna3.scaffold_13337 15591038 54 + 23293914 ----------UACCCGUGGAUUAAGACUCAUAAAAUGCGGGAGAGCAGAG--UGGGAUUUACCCAA ----------..((((((..(((.......)))..)))))).........--((((.....)))). ( -12.60, z-score = -1.11, R) >droSim1.chr3L 8384598 64 - 22553184 UUUUUUGCCAAUCUU-UAAAGAAGAGCGGCAACACUCC-CAUGAGAAGAAUUUGGGAUUUACCCAA ....(((((..((((-(...)))))..)))))..(((.-...)))......(((((.....))))) ( -15.40, z-score = -2.18, R) >droSec1.super_0 1218007 64 - 21120651 UUUUUUGCCAAUCUU-UAAAGAAGAGCGGCAACACUCC-CAUGAGAAGAAUUUGGGAUUUACCCAA ....(((((..((((-(...)))))..)))))..(((.-...)))......(((((.....))))) ( -15.40, z-score = -2.18, R) >droWil1.scaffold_180697 3837853 66 - 4168966 AACAUUGCCAUUAUUACCAGGAAAAGACCAGAGAAUACAUCGAAGUAGCAUUUUAGUUUAAGCAAA ....((((....((((...((......))......(((......)))......))))....)))). ( -4.70, z-score = 0.01, R) >consensus UUUUUUGCCAAUCUU_UAAAGAAGAGCGACAAAACUAC_CAUAAGAAGAAUUUGGGAUUUACCCAA ....................................................((((.....)))). ( -3.60 = -3.64 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:12:40 2011