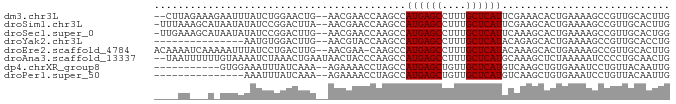
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,905,233 – 8,905,315 |
| Length | 82 |
| Max. P | 0.903526 |

| Location | 8,905,233 – 8,905,315 |
|---|---|
| Length | 82 |
| Sequences | 8 |
| Columns | 86 |
| Reading direction | forward |
| Mean pairwise identity | 66.19 |
| Shannon entropy | 0.67509 |
| G+C content | 0.41245 |
| Mean single sequence MFE | -18.14 |
| Consensus MFE | -8.90 |
| Energy contribution | -8.79 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -0.74 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.723672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
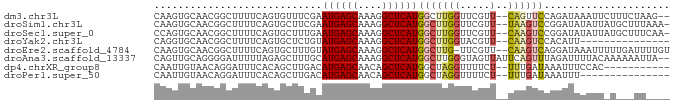
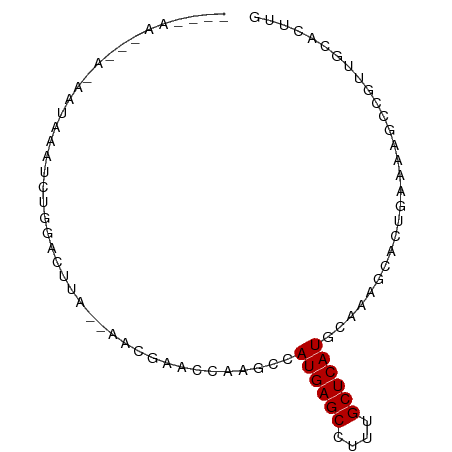
>dm3.chr3L 8905233 82 + 24543557 CAAGUGCAACGGCUUUUCAGUGUUUCGAAUGAGCAAAGGCUCAUGGCUUGGUUCGUU--CAGUUCCAGAUAAAUUCUUUCUAAG-- ....((.(((.(((....))).....((((((((.(((.(....).))).)))))))--).))).)).................-- ( -15.60, z-score = 0.30, R) >droSim1.chr3L 8340934 83 + 22553184 CAAGUGCAACGGCUUUUCAGUGCUUCGAAUGAGCAAAGGCUCAUGGCUUGGUUCGUU--UAAGUCCGGAUAUAUUAUGCUUUAAA- .....(((...((........))((((.((((((....))))))(((((((.....)--)))))))))).......)))......- ( -18.10, z-score = 0.14, R) >droSec1.super_0 1175761 83 + 21120651 CCAGUGCAACGGCUUUUCAGUGCUUUGAAUGAGCAAAGGCUCAUGGCUUGGUUCGUU--CAAGUCCGGAUAUAUUAUGCUUUCAA- .....(((..(((........)))((((((((((.(((.(....).))).)))))))--)))..............)))......- ( -20.00, z-score = -0.08, R) >droYak2.chr3L 21188572 69 - 24197627 CAGGUGCAACGGCUUUUCAGUGCUCUGUAUGAGCAAAGGCUCAUGGCUUGGUACGUU--CAAGUCCACAUU--------------- ...(((....(((((....(((((..((((((((....)))))).))..)))))...--.))))))))...--------------- ( -21.00, z-score = -1.44, R) >droEre2.scaffold_4784 20623615 83 - 25762168 CAAGUGCAACGGCUUUUCAGUGCUUUGUAUGAGCAAAGGCUCAUGGCUUG-UUCGUU--CAAGUCAGGAUAAAUUUUUGAUUUUGU (((((((((.(((........)))))))((((((....)))))).)))))-......--.(((((((((.....)))))))))... ( -21.30, z-score = -1.16, R) >droAna3.scaffold_13337 15549265 84 - 23293914 CAGUUGCAGGGGAUUUUUAGAGCUUUGCAUGAGCAAAGGCUCAUGGCUUGGGUAGUUAUUCAGUUUAGAUUUUACAAAAAAUUA-- ...........(((((((.((((....(((((((....)))))))))))..((((....((......))..)))).))))))).-- ( -16.50, z-score = -0.15, R) >dp4.chrXR_group8 557131 73 - 9212921 CAAUUGUAACAGGAUUUCACAGCUUGACAUGAGCAACAGCUCAUGGCUAGGUUUUCU--UUUGAUAAAUUUCCAC----------- ...........(((......((((((.(((((((....)))))))..)))))).((.--...))......)))..----------- ( -16.80, z-score = -1.83, R) >droPer1.super_50 543442 69 + 557249 CAAUUGUAACAGGAUUUCACAGCUUGACAUGAGCAACAGCUCAUGGCUAGGUUUUCU--UUUGAUAAAUUU--------------- ...((((...((((......((((((.(((((((....)))))))..)))))).)))--)...))))....--------------- ( -15.80, z-score = -1.74, R) >consensus CAAGUGCAACGGCUUUUCAGUGCUUUGAAUGAGCAAAGGCUCAUGGCUUGGUUCGUU__CAAGUCCAGAUAUAUU_U___UU____ .....................(((....((((((....)))))))))....................................... ( -8.90 = -8.79 + -0.11)
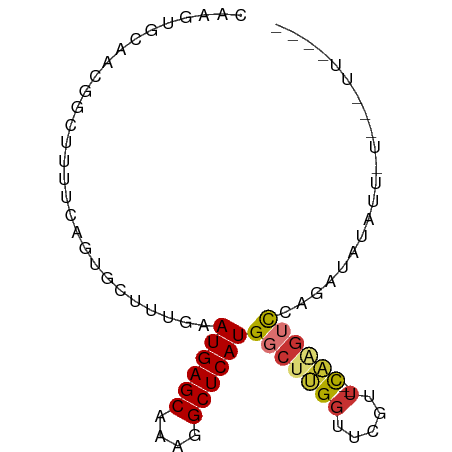
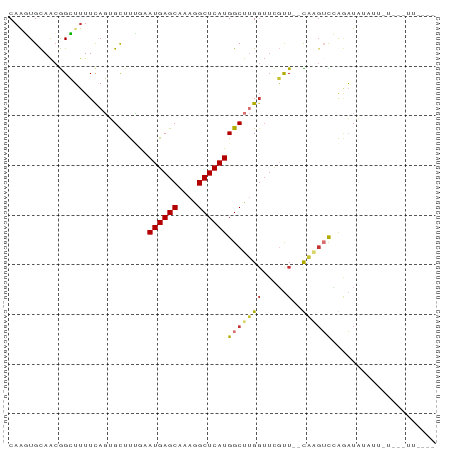
| Location | 8,905,233 – 8,905,315 |
|---|---|
| Length | 82 |
| Sequences | 8 |
| Columns | 86 |
| Reading direction | reverse |
| Mean pairwise identity | 66.19 |
| Shannon entropy | 0.67509 |
| G+C content | 0.41245 |
| Mean single sequence MFE | -13.66 |
| Consensus MFE | -6.88 |
| Energy contribution | -6.88 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.903526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8905233 82 - 24543557 --CUUAGAAAGAAUUUAUCUGGAACUG--AACGAACCAAGCCAUGAGCCUUUGCUCAUUCGAAACACUGAAAAGCCGUUGCACUUG --.((((..(((.....)))....)))--)......((((..((((((....))))))..(.(((.((....))..))).).)))) ( -11.00, z-score = 0.51, R) >droSim1.chr3L 8340934 83 - 22553184 -UUUAAAGCAUAAUAUAUCCGGACUUA--AACGAACCAAGCCAUGAGCCUUUGCUCAUUCGAAGCACUGAAAAGCCGUUGCACUUG -......(((.........(((.(((.--..............(((((....)))))((((......)))))))))).)))..... ( -12.10, z-score = 0.10, R) >droSec1.super_0 1175761 83 - 21120651 -UUGAAAGCAUAAUAUAUCCGGACUUG--AACGAACCAAGCCAUGAGCCUUUGCUCAUUCAAAGCACUGAAAAGCCGUUGCACUGG -(((((..............((.((((--.......)))))).(((((....)))))))))).(((.((......)).)))..... ( -17.00, z-score = -0.91, R) >droYak2.chr3L 21188572 69 + 24197627 ---------------AAUGUGGACUUG--AACGUACCAAGCCAUGAGCCUUUGCUCAUACAGAGCACUGAAAAGCCGUUGCACCUG ---------------...((((.((((--.......))))))))((((....))))...(((.(((.((......)).)))..))) ( -16.10, z-score = -1.08, R) >droEre2.scaffold_4784 20623615 83 + 25762168 ACAAAAUCAAAAAUUUAUCCUGACUUG--AACGAA-CAAGCCAUGAGCCUUUGCUCAUACAAAGCACUGAAAAGCCGUUGCACUUG .....................(.((((--......-)))).)((((((....)))))).....(((.((......)).)))..... ( -12.10, z-score = -0.70, R) >droAna3.scaffold_13337 15549265 84 + 23293914 --UAAUUUUUUGUAAAAUCUAAACUGAAUAACUACCCAAGCCAUGAGCCUUUGCUCAUGCAAAGCUCUAAAAAUCCCCUGCAACUG --.......(((((...((......)).................((((.(((((....)))))))))...........)))))... ( -13.20, z-score = -2.97, R) >dp4.chrXR_group8 557131 73 + 9212921 -----------GUGGAAAUUUAUCAAA--AGAAAACCUAGCCAUGAGCUGUUGCUCAUGUCAAGCUGUGAAAUCCUGUUACAAUUG -----------..(((......((...--.))...(.(((((((((((....)))))))....)))).)...)))........... ( -14.80, z-score = -1.40, R) >droPer1.super_50 543442 69 - 557249 ---------------AAAUUUAUCAAA--AGAAAACCUAGCCAUGAGCUGUUGCUCAUGUCAAGCUGUGAAAUCCUGUUACAAUUG ---------------............--......(.(((((((((((....)))))))....)))).)................. ( -13.00, z-score = -1.73, R) >consensus ____AA___A_AAUAAAUCUGGACUUA__AACGAACCAAGCCAUGAGCCUUUGCUCAUGCAAAGCACUGAAAAGCCGUUGCACUUG ..........................................((((((....))))))............................ ( -6.88 = -6.88 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:12:36 2011