| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,888,369 – 8,888,459 |
| Length | 90 |
| Max. P | 0.822485 |

| Location | 8,888,369 – 8,888,459 |
|---|---|
| Length | 90 |
| Sequences | 13 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 66.64 |
| Shannon entropy | 0.72846 |
| G+C content | 0.40534 |
| Mean single sequence MFE | -17.92 |
| Consensus MFE | -8.49 |
| Energy contribution | -8.00 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.822485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8888369 90 - 24543557 ----------AUUACAUUU-CCUUCAAUUGAUAUUAAAUUUAUU-AUGUCAGGUGUCUUUAUGAUCCUUAUGCCCAUUCUUCUGAUGAACUUGCUCAUCGGU ----------....(((..-....((.(((((((..........-))))))).)).....)))..................(((((((......))))))). ( -13.00, z-score = -0.55, R) >droSim1.chr3L 8324096 90 - 22553184 ----------AUUACUUUU-CCUUUAAUUAAUAUCAAACUUAUU-AUGUCAGGUGUCUUUAUGAUCCUUAUGCCCAUUCUGCUGAUGAACUUGCUCAUCGGU ----------.........-........(((((.......))))-).((.(((.(((.....)))))).)).........((((((((......)))))))) ( -12.30, z-score = -0.71, R) >droSec1.super_0 1159018 90 - 21120651 ----------AUUACUUUU-CCUUUAAUUGAUAUUAAAUUUAUU-AUGCCAGGUGUCUUUAUGAUCCUUAUGCCCAUUCUUCUGAUGAACUUGCUCAUCGGU ----------.........-..((((((....))))))......-..((.(((.(((.....))))))...))........(((((((......))))))). ( -11.90, z-score = -0.54, R) >droYak2.chr3L 21172281 91 + 24197627 ----------AUUACUUUUCCCUUUAACACAUAUUAUGUGUUUG-CUGGCAGGCGUCUUUAUGAUCCUUAUGCCCAUUCUGCUGAUGAACUUGCUCAUCGGU ----------...............(((((((...)))))))..-..((((((.(((.....)))))...))))......((((((((......)))))))) ( -20.10, z-score = -1.93, R) >droEre2.scaffold_4784 20607192 90 + 25762168 ----------AUUGCUUUU-CCUUUAACAGCUAUCAUGUGUUUG-AUGGCAGGCGUCUUUAUGAUCCUCAUGCCCAUUCUCCUGAUGAACUUGCUCAUCGGU ----------...((....-.........(((((((......))-)))))(((.(((.....))))))...))........(((((((......))))))). ( -21.20, z-score = -1.37, R) >droAna3.scaffold_13337 15532138 94 + 23293914 --------AAGAUACUCCUUCCAGGAUACGAUAAAAUCUUAUCCUUUUUAAGGUGUGUUUAUGAUCCUCAUGCCCAUUCUCCUGAUGAACUUGCUCAUCGGU --------.((((((.((((..((((((.(((...))).))))))....)))).))))))(((.....)))..........(((((((......))))))). ( -23.20, z-score = -2.77, R) >dp4.chrXR_group8 539981 92 + 9212921 --------AUUCCCACCUGUCCAUGAUCUGAUUGCUAUUUCUCG-UG-ACAGGCGUCUUCAUGAUACUCAUGCCCAUUCUGUUGAUGAAUUUGCUUAUCGGU --------....((.((((((.((((...(((....)))..)))-))-)))))......((((.....))))...........(((((......))))))). ( -17.20, z-score = 0.01, R) >droPer1.super_50 526093 92 - 557249 --------AUUCCCACCUGUCCAUGAUCUGAUGUCUAUUUCUCG-UG-ACAGGCGUCUUCAUGAUACUCAUGCCCAUUCUGUUGAUGAAUUUGCUUAUCGGU --------......(((....(((((...(((((((.((.....-.)-).))))))).)))))........((..((((.......))))..)).....))) ( -18.60, z-score = -0.42, R) >droWil1.scaffold_180698 3502334 90 + 11422946 ----------GCUCCACCU-CCUUCGGCUC-UCUUAUGUUUGUGUCUUGCAGGCGUCUUUAUGAUCCUCAUGCCCAUUCUAUUGAUGAAUCUGCUUAUCGGU ----------.....(((.-.....(((..-....((((((((.....))))))))....(((.....)))))).........(((((......)))))))) ( -15.20, z-score = -0.08, R) >droVir3.scaffold_13049 6574592 92 - 25233164 --------GUACCC-UCUAGCAAUCUGCUAAUCUCUAUAAUUUA-ACGACAGGUGUGUUUAUGAUUCUGAUGCCCAUUCUGUUGAUGAACUUGCUCAUCGGU --------..(((.-..((((.....))))..............-..((((((.(((..(((.......)))..)))))))))(((((......)))))))) ( -15.70, z-score = -0.14, R) >droMoj3.scaffold_6680 4894310 95 - 24764193 -AUAGUCUGUCCAC-CCUUUCCCU-UGCUAAUCUGCA-AAUCU---CUUCAGGUGUCUUUAUGAUUCUGAUGCCCAUUCUGCUGAUGAACUUGCUCAUCGGU -..((..((..((.-........(-(((......)))-)....---..(((((.(((.....))))))))))..))..))((((((((......)))))))) ( -19.10, z-score = -1.67, R) >droGri2.scaffold_15110 18367045 91 + 24565398 ---------AAACACUUUAACUAGG-CACG-UCUAAUCAUCUUUCCUGGCAGGUGUUUUUAUGGUCUUGAUGCCCAUUCUGUUGAUGAACUUGCUCAUCGGU ---------((((((((...(((((-....-.............))))).))))))))..((((.(.....).))))....(((((((......))))))). ( -16.73, z-score = 0.34, R) >anoGam1.chr2L 44735261 100 + 48795086 GCAGGCAAGUGUGCAGACGUACACGGGCUAAU-GCAAUUUUUGU-UGCGCAGGUUUGUUCAUGAUCCUUAUGCCGAUUCUGUUGAUGAAUCUGUUGAUCGGU ...(((..((((((....))))))..)))...-(((((....))-)))(((((((..((((.((((........)))).))..))..)))))))........ ( -28.70, z-score = -0.35, R) >consensus __________AUUACUCUU_CCAUGAACUGAUAUCAAUUUUUUG_AUGGCAGGUGUCUUUAUGAUCCUCAUGCCCAUUCUGCUGAUGAACUUGCUCAUCGGU ...................................................(((((.............))))).......(((((((......))))))). ( -8.49 = -8.00 + -0.49)

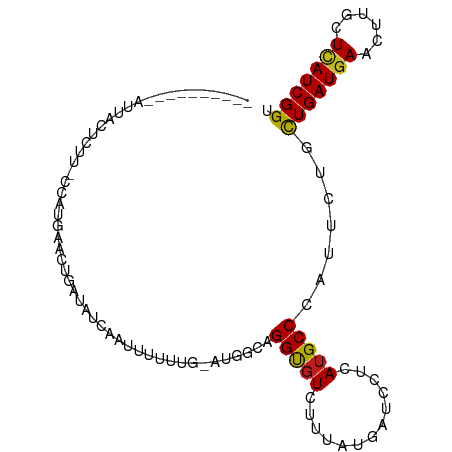

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:12:34 2011