
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,872,597 – 8,872,691 |
| Length | 94 |
| Max. P | 0.887861 |

| Location | 8,872,597 – 8,872,691 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 68.88 |
| Shannon entropy | 0.62908 |
| G+C content | 0.42550 |
| Mean single sequence MFE | -22.17 |
| Consensus MFE | -9.96 |
| Energy contribution | -9.99 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.887861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
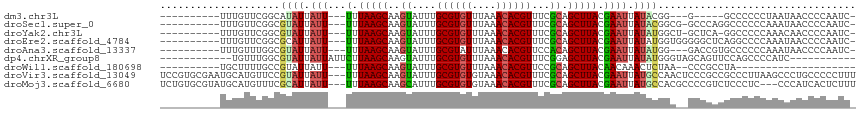
>dm3.chr3L 8872597 94 + 24543557 ----------UUUGUUCGGCAUAUUAUU---UUUAAGCAAGUAUUUGCGUGUUUAAACACGUUUCGCAGCUUACGAAUUAUACGG---G-----GCCCCCCUAAUAACCCCAAUC- ----------.......((..(((.(((---(.(((((..((....((((((....))))))...)).))))).)))).))).((---(-----....)))........))....- ( -19.80, z-score = -0.99, R) >droSec1.super_0 1143257 101 + 21120651 ----------UUUGUUCGGCGUAUUAUU---UUUAAGCAAGUAUUUGCGUGUUUAAACACGUUUCGCAGCUUACGAAUUAUACGGCG-GCCCAGGCCCCCCAAAUAACCCCAAUC- ----------.(((((.((.((((.(((---(.(((((..((....((((((....))))))...)).))))).)))).))))((.(-((....))))))).)))))........- ( -27.50, z-score = -2.36, R) >droYak2.chr3L 21156206 100 - 24197627 ----------UUUGUUCGGCGUAUUAUU---UUUAAGCAAGUAUUUGCGUGUUUAAACACGUUUCGCAGCUUACGAAUUAUAUGGCU-GCUCA-GGCCCCCAAACAACCCCAAUC- ----------.(((((.((.((((.(((---(.(((((..((....((((((....))))))...)).))))).)))).))))((((-.....-)))).)).)))))........- ( -24.00, z-score = -1.78, R) >droEre2.scaffold_4784 20591408 102 - 25762168 ----------UUUGUUCGGCGCAUUAUU---UUUAAGCAAGUAUUUGCGUGUUUAAACACGUUUCGCAGCUUACGAAUUAUAUGGUGGGGGCUCAGGCCCCAAAUAACCCCAAUC- ----------(((((..(((((.(((..---..)))))..((....((((((....))))))...)).))).)))))......(((.(((((....))))).....)))......- ( -27.30, z-score = -1.30, R) >droAna3.scaffold_13337 15518602 99 - 23293914 ----------UUUGUUUGGCGUAUUAUU---UUUAAGCAAGUAUUUGCGUAUUUAAACACGUUCCACAGCUUACGAAUUAUAUGG---GACCGUGCCCCCCAAAUAACCCCAAUC- ----------.((((((((.((((.(((---(.(((((..((....((((........))))...)).))))).)))).))))((---(......))).))))))))........- ( -20.10, z-score = -1.28, R) >dp4.chrXR_group8 522613 93 - 9212921 ------------UGUUUGGCGUAUUAUUAUUCUUAAGCAAGUAUUUGCGUGUUUAAACACGUUUCGGAGCUUACGAAUUAUAUGGGUAGCAGUUCCAGCCCCAUC----------- ------------.....(((....(((.((((.(((((........((((((....))))))......))))).)))).)))((((.......))))))).....----------- ( -21.64, z-score = -1.17, R) >droWil1.scaffold_180698 3480956 81 - 11422946 ----------UGCUUUUGCCGUAUUAUU---UUUAAGCAAGUAUUUGCGUGUUUAAACACGUUCCGCAGCUUACAACAAACUCUAA--CCCGCCUA-------------------- ----------.((....)).........---..(((((..((....((((((....))))))...)).))))).............--........-------------------- ( -11.30, z-score = 0.15, R) >droVir3.scaffold_13049 6558016 113 + 25233164 UCCGUGCGAAUGCAUGUUCCGUAUUAUU---UUUAAGCAAGUAUUUGCGUGUGUAAACACGUUUCGCAGCUUACGAAUUAUGCCAACUCCCGCCGCCCUUAAGCCCUGCCCCCUUU ..(((((....)))))....((((.(((---(.(((((..((....((((((....))))))...)).))))).)))).))))........((.((......))...))....... ( -22.60, z-score = -1.38, R) >droMoj3.scaffold_6680 4878188 110 + 24764193 UCUGUGCGUAUGCAUGUUUCGCAUUAUU---UUUAAGCAAGCAUUUGCGUGUGUAAACACGUUUCGCAGCUUACGAAUUAUGCCACGCCCCGUCUCCCUC---CCCAUCACUCUUU .....((((..(((((.((((.......---..(((((..((....((((((....))))))...)).))))))))).))))).))))............---............. ( -25.30, z-score = -2.59, R) >consensus __________UUUGUUUGGCGUAUUAUU___UUUAAGCAAGUAUUUGCGUGUUUAAACACGUUUCGCAGCUUACGAAUUAUACGGC__GCCCUCGCCCCCCAAACAACCCCAAUC_ .................................(((((..((....((((((....))))))...)).)))))........................................... ( -9.96 = -9.99 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:12:31 2011