| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,822,589 – 8,822,691 |
| Length | 102 |
| Max. P | 0.972259 |

| Location | 8,822,589 – 8,822,691 |
|---|---|
| Length | 102 |
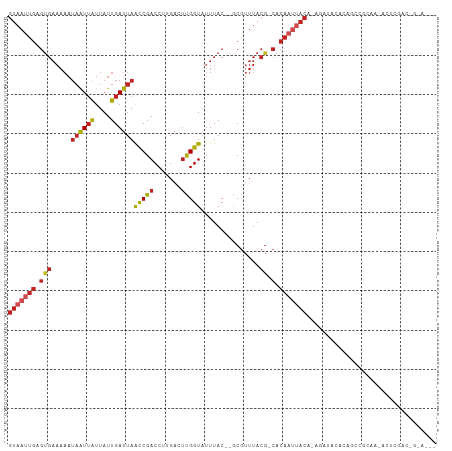
| Sequences | 11 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 76.62 |
| Shannon entropy | 0.49765 |
| G+C content | 0.36331 |
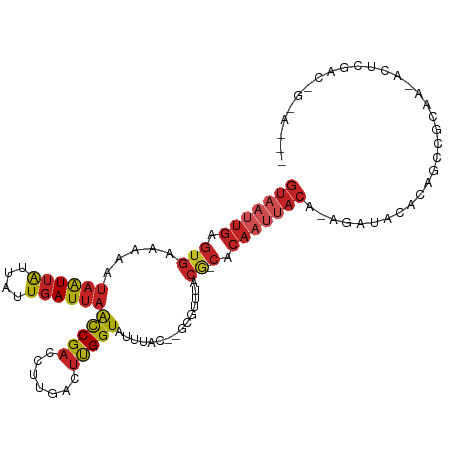
| Mean single sequence MFE | -19.55 |
| Consensus MFE | -9.32 |
| Energy contribution | -10.73 |
| Covariance contribution | 1.41 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.972259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8822589 102 + 24543557 GUAAUUGAGUGAAAAAUAAUUAUUAUUGAUUAGCCGACCUUGACUUGGUAUUUAC--GCGUUUACG-CACAAUUACA-UGAUACACAGCCGCAA-ACUCGACUGGAA-- (((((((.(((((...((((((....))))))(((((.......))))).)))))--(((....))-).))))))).-.......(((.((...-...)).)))...-- ( -22.70, z-score = -2.38, R) >droSim1.chr3L 8261728 102 + 22553184 GUAAUUGAGUGAAAAAUAAUUAUUAUUGAUUAACCGACCUUGACUUGGUAUUUAC--GCGUUUACG-CACAAUUACA-UGAUACACAGCCGCAA-ACUCGACUGGAA-- (((((((.(((((...((((((....))))))(((((.......))))).)))))--(((....))-).))))))).-.......(((.((...-...)).)))...-- ( -22.40, z-score = -2.76, R) >droSec1.super_0 1093400 84 + 21120651 ------------------GUAAUUAUUGAUUAACCGACCUUGACUUGGUAUUUAC--GCGUUUACG-CACAAUUACA-UGAUACACUGACGCAA-ACUCGACUGGAA-- ------------------((((((........(((((.......)))))......--(((....))-)..)))))).-................-............-- ( -11.10, z-score = -0.17, R) >droYak2.chr3L 21105987 102 - 24197627 GUAAUUGAGUGAAAAAUAAUUAUUAUUGAUUAACCGACCUUGACUUGGUAUUUAC--GCGUUUACG-CACAAUUACA-UGAUACACAGCCGCAA-ACUCGACUGGAA-- (((((((.(((((...((((((....))))))(((((.......))))).)))))--(((....))-).))))))).-.......(((.((...-...)).)))...-- ( -22.40, z-score = -2.76, R) >droEre2.scaffold_4784 20541104 102 - 25762168 GUAAUUGAGUGAAAAAUAAUUAUUAUUGAUUAACCGACCUUGACUUGGUAUUUAC--GCGUUUACG-CACAAUUACA-UGAUACACAGCCGCAA-ACUCGACUGGAA-- (((((((.(((((...((((((....))))))(((((.......))))).)))))--(((....))-).))))))).-.......(((.((...-...)).)))...-- ( -22.40, z-score = -2.76, R) >droAna3.scaffold_13337 15471615 105 - 23293914 GUAAUUGAGUGAAAAAUAAUUAUUAUUGAUUAACCGACCUUGACUCGGUAUUUAC--GCGUUUACG-CACAAUUACA-AGAUACAGAUCCCCACCAGACUGCCGCAGAA (((((((.(((((...((((((....))))))(((((.......))))).)))))--(((....))-).))))))).-.(...(((.((.......)))))...).... ( -22.50, z-score = -3.27, R) >dp4.chrXR_group8 455584 102 - 9212921 GUAAUUGAGUGAAAAAUAAUUAUUAUUGAUUAACCGACCUUGACUUGGUCUUUACGCACAAUUACAACACACGCACACACACACACGCAUGCACCCGUGAGA------- (((((((.(((.....((((((....))))))...((((.......))))....))).)))))))..................((((........))))...------- ( -21.30, z-score = -2.69, R) >droWil1.scaffold_180698 3412821 83 - 11422946 GUAAUUGAGUGAAAAAUAAUUGUUAUUGAUUAAUCGACCUUGAUGUGGUAUUUAC--ACAUUUACG-CACAAUUACA-AGACACCCA---------------------- (((((((.(((.....(((((......))))).........((((((.......)--)))))..))-).))))))).-.........---------------------- ( -16.30, z-score = -1.88, R) >droVir3.scaffold_13049 6499190 92 + 25233164 GUAAUUGAGUGAAAAAUAAUUAUUAUUGAUUAACCGACCUUGACUUGGUAUUUAC--ACGUUUACG-CACAAUUACG-AGGCA-GCAGCAGCAGCAG------------ (((((((.(((.....((((((....))))))(((((.......)))))......--.......))-).))))))).-..((.-((....)).))..------------ ( -18.70, z-score = -1.46, R) >droMoj3.scaffold_6680 4821897 77 + 24764193 GUAAUUGAGUGAAAAAUAAUUAUUAUUGAUUAACCGACCUUGACUUGGUAUUUAC--ACGUUUACG-CACAAUUACA-AAG---------------------------- (((((((.(((.....((((((....))))))(((((.......)))))......--.......))-).))))))).-...---------------------------- ( -15.40, z-score = -2.90, R) >droGri2.scaffold_15110 18303116 95 - 24565398 GUAAUUGAGUGAAAAAUAAUUAUUAUUGAUUAACCGACCUUGACUUGGUAUUUAC--ACGUUUACG-CACAAUUACA-AGGCA-GCAGCCAAAGCAGAAG--------- (((((((.(((.....((((((....))))))(((((.......)))))......--.......))-).))))))).-.(((.-...)))..........--------- ( -19.90, z-score = -2.38, R) >consensus GUAAUUGAGUGAAAAAUAAUUAUUAUUGAUUAACCGACCUUGACUUGGUAUUUAC__GCGUUUACG_CACAAUUACA_AGAUACACAGCCGCAA_ACUCGAC_G_A___ (((((((.(((((...((((((....))))))(((((.......)))))...........)))))....)))))))................................. ( -9.32 = -10.73 + 1.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:12:26 2011