
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,634,757 – 4,634,847 |
| Length | 90 |
| Max. P | 0.716226 |

| Location | 4,634,757 – 4,634,847 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 86.28 |
| Shannon entropy | 0.26773 |
| G+C content | 0.43628 |
| Mean single sequence MFE | -18.52 |
| Consensus MFE | -14.96 |
| Energy contribution | -15.16 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.716226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4634757 90 - 23011544 --CUUUUUUGCCC----CCAUGCCUUGUCCAUGCUUAUUUUCUAUUACUGCUUUUUCCACUUAAGGCCAUUGCACUUUUUACGUGC-AUGCCAUGGA --...........----..........(((((((...............(((((........)))))...(((((.......))))-).).)))))) ( -15.50, z-score = -1.35, R) >droSec1.super_5 2719850 90 - 5866729 --CUUUUUUGCCC----CAAUGCCUUGUCCAUGCUUAUUUUCUAUUAGUGCUUUUUCCACUUAAGGCCAUUGCACUUUUUACGUGC-AUGCCAUGGA --...........----..........(((((..............((((.......))))...(((...(((((.......))))-).)))))))) ( -16.70, z-score = -1.12, R) >droYak2.chr2L 4649726 90 - 22324452 --CUUUUUUGCCC----CCAUGCCUUGUCCAUGCUUAUUUUCUAUUACUGCUUUUUCCACUUAAGGCCAUUGCACUUUUUACGUGC-AUGCCAUGGA --...........----..........(((((((...............(((((........)))))...(((((.......))))-).).)))))) ( -15.50, z-score = -1.35, R) >droEre2.scaffold_4929 4715700 90 - 26641161 --CUUUUUUGCCC----CCAUGCCUUGUCCAUGCUUAUUUUCUAUUACUGCUUUUUCCACUUAAGGCCAUUGCACUUUUUACGUGC-AUGCCAUGGA --...........----..........(((((((...............(((((........)))))...(((((.......))))-).).)))))) ( -15.50, z-score = -1.35, R) >dp4.Unknown_group_675 19299 94 - 20316 --CCUUUUGGCCCCAUGCCAUGCCUGGUCCAUGCUUAUUUUCUAUUACUGCUUUUUCCACUUAAGGCCAUUGCACUUUUUACGUGC-AUGCCACGGA --.....((((.....))))..(((((.....((...............)).....))).....(((...(((((.......))))-).)))..)). ( -19.86, z-score = -0.71, R) >droPer1.super_8 1528573 94 + 3966273 --CCUUUUGGCCCCAUGCCAUGCCUGGUCCAUGCUUAUUUUCUAUUACUGCUUUUUCCACUUAAGGCCAUUGCACUUUUUACGUGC-AUGCCACGGA --.....((((.....))))..(((((.....((...............)).....))).....(((...(((((.......))))-).)))..)). ( -19.86, z-score = -0.71, R) >droWil1.scaffold_180703 1376378 92 + 3946847 UUCUCUUCUUGCC----AGGCCUCUGGUCCAUGCUUAUUUUCUAUUACUGCUUUUUCCACUUAAGGCCAUUGCACUUUUUACGUGC-AUGCCAUGGA ...........((----((((((.(((.....((...............)).....)))....)))))..(((((.......))))-).....))). ( -20.36, z-score = -1.75, R) >droMoj3.scaffold_6500 12010170 87 + 32352404 ------UUGCUGC----AGCCCCAGAGUCCAUGCUUAUUUUCUAUUACUGCUUUUUCCACUUAAGGCCAUUGCACUUUUUACGUGCAAUGCCAUGGA ------...(((.----.....)))..((((((................(((((........)))))((((((((.......)))))))).)))))) ( -20.70, z-score = -1.58, R) >droVir3.scaffold_12963 8547398 85 + 20206255 --------GCUCC----AGCCCCAGAGUCCAUGCUUAUUUUCUAUUACUGCUUUUUCCACUUAAGGCCAUUGCACUUUUUACGUGCAAUGCCAUGGA --------((((.----.......))))(((((................(((((........)))))((((((((.......)))))))).))))). ( -21.00, z-score = -2.67, R) >droGri2.scaffold_15126 3185259 86 + 8399593 -------UGCUAC----AGCCCCAGAGUCCAUGCUUAUUUUCUAUUACUGCUUUUUCCACUUAAGGCCAUUGCACUUUUUACGUGCAAUGCCAUGGA -------.((...----.)).......((((((................(((((........)))))((((((((.......)))))))).)))))) ( -20.20, z-score = -2.00, R) >consensus __CUUUUUGGCCC____CCAUGCCUUGUCCAUGCUUAUUUUCUAUUACUGCUUUUUCCACUUAAGGCCAUUGCACUUUUUACGUGC_AUGCCAUGGA ...........................((((((................(((((........)))))(((.((((.......)))).))).)))))) (-14.96 = -15.16 + 0.20)

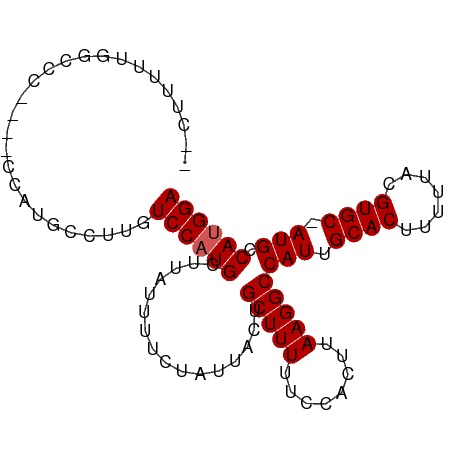
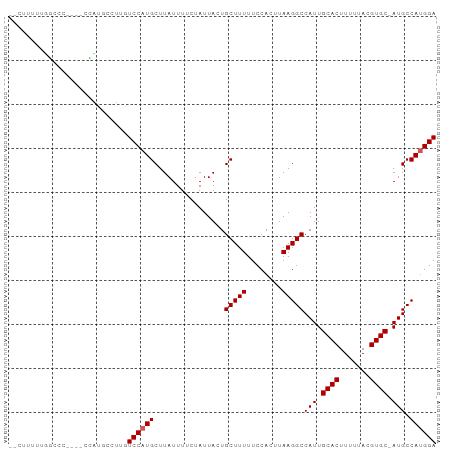
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:17:02 2011