| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,729,766 – 8,729,898 |
| Length | 132 |
| Max. P | 0.807017 |

| Location | 8,729,766 – 8,729,898 |
|---|---|
| Length | 132 |
| Sequences | 7 |
| Columns | 146 |
| Reading direction | reverse |
| Mean pairwise identity | 55.41 |
| Shannon entropy | 0.85070 |
| G+C content | 0.33217 |
| Mean single sequence MFE | -25.59 |
| Consensus MFE | -6.66 |
| Energy contribution | -8.11 |
| Covariance contribution | 1.46 |
| Combinations/Pair | 1.68 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.807017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
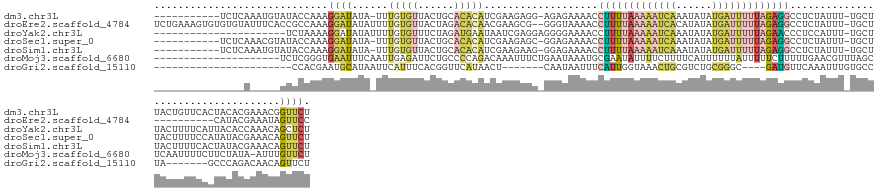
>dm3.chr3L 8729766 132 - 24543557 -----------UCUCAAAUGUAUACCAAAGGAUAUA-UUUGUGUUACUGCACACAUCGAAGAGG-AGAGAAAACCUUUUAAAAAUCAAAUAUAUGAUUUUUAGAGGCCUCUAUUU-UGCUUACUGUUCACUACACGAAACGGUUCU -----------((((((((((((.((...)))))))-)))((((....))))........))))-((((.....(((((((((((((......))))))))))))).))))....-.....(((((((.......).))))))... ( -27.70, z-score = -1.79, R) >droEre2.scaffold_4784 20448971 133 + 25762168 UCUGAAAGUGUGUGUAUUUCACCGCCAAAGGAUAUAUUUUGUGUUACUAGACACAACGAAGCG--GGGUAAAACCUUUUAAAAAUCACAUAUAUGAUUUUUAGAGGCCUCUAUUU-UGCU----------CAUACGAAAUAGUUCC .......(((.(((.....))))))....(((..(((((((((((....)))))...((.(((--(((((....(((((((((((((......)))))))))))))....)))))-))))----------)....))))))..))) ( -32.60, z-score = -2.34, R) >droYak2.chr3L 21016954 123 + 24197627 ----------------------UCUAAAAGGAUAUAUUUUGUGUUUCUAGAUGAAUAAUCGAGGAGGGGAAAACCUUUUAAAAAUCAAAUAUAUGAUUUUUAGAACCCUCCAUUU-UGCUUACUUUUCAUUACACCAAACAGCUCU ----------------------.......(((....(((.((((.....(((((((((.(((((((((.......((((((((((((......))))))))))))))))))...)-)).)))...)))))))))).)))....))) ( -24.01, z-score = -1.40, R) >droSec1.super_0 1007421 132 - 21120651 -----------UCUCAAACGUAUACCAAAGGAUAUA-UUUGUGUUACUGCACACAUCGAAGAGC-GGAGAAAACCUUUUAAAAAUCAAAUAUAUGAUUUUUAGAGGCCUCUAUUU-UGCUUACUUUUCCAUAUACGAAACAGUUCU -----------.......((((((..(((((.....-..(((((....))))).......((((-((((.....(((((((((((((......)))))))))))))......)))-))))).)))))...)))))).......... ( -29.30, z-score = -3.08, R) >droSim1.chr3L 8173388 132 - 22553184 -----------UCUCAAAUGUAUACCAAAGGAUAUA-UUUGUGUUACUGCACACAUCGAAGAAG-GGAGAAAACCUUUUAAAAAUCAAAUAUAUGAUUUUUAGAGGCCUCUAUUU-UGCUUACUUUUCACUAUACGAAACAGUUCU -----------.......((((((..(((((.....-..(((((....)))))...(((((...-((((.....(((((((((((((......))))))))))))).)))).)))-))....)))))...)))))).......... ( -24.30, z-score = -1.22, R) >droMoj3.scaffold_6680 4712854 124 - 24764193 ---------------------UCUCGGGUGAAUUUCAAUUGAGAUUCUGCCCCAGACAAAUUUCUGAAUAAAUGCGAAUAUUUUCUUUUCAUUUUUUAUUUUUCUUUUUGAACGUUUAGCUCAAUUUUCUUCUAUA-AUUUGUUCU ---------------------(((.(((((((((((....))))))).)))).))).........((((((((..(((....)))......................((((.(.....).))))............-)))))))). ( -22.20, z-score = -1.87, R) >droGri2.scaffold_15110 18214228 105 + 24565398 -----------------------CCACGAAUGCAUAAUUCAUUUCACGGUUCAUAACU-------CAAUAAUUUCAUUGGUAAACUGCGUCUGCGGGC----GAUGUUCAAAUUUGUGCCUA-------GCCCAGACAACAGUUCU -----------------------....((((.....))))...............((.-------((((......)))))).(((((.((((..((((----...((.((....)).))...-------))))))))..))))).. ( -19.00, z-score = 0.39, R) >consensus _______________A_____AUACCAAAGGAUAUA_UUUGUGUUACUGCACACAUCGAAGAGG_GGAGAAAACCUUUUAAAAAUCAAAUAUAUGAUUUUUAGAGGCCUCUAUUU_UGCUUACUUUUCACUAUACGAAACAGUUCU .............................((((......(((((......)))))...................(((((((((((((......)))))))))))))...................................)))). ( -6.66 = -8.11 + 1.46)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:12:20 2011