| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,710,474 – 8,710,573 |
| Length | 99 |
| Max. P | 0.991377 |

| Location | 8,710,474 – 8,710,573 |
|---|---|
| Length | 99 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 63.86 |
| Shannon entropy | 0.70014 |
| G+C content | 0.47204 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -12.40 |
| Energy contribution | -12.41 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.991377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
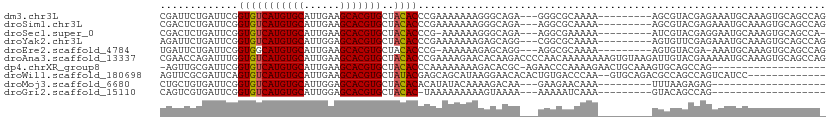
>dm3.chr3L 8710474 99 - 24543557 CGAUUCUGAUUCGGUGUCAUGUGCAUUGAAGCACGUGCUACACCCGAAAAAAAGGGCAGA---GGGCGCAAAA---------AGCGUACGAGAAAUGCAAAGUGCAGCCAG ...(((((.....(((.(((((((......))))))).))).(((........)))))))---)((((((...---------.((((.......))))....))).))).. ( -32.50, z-score = -2.27, R) >droSim1.chr3L 8153312 99 - 22553184 CGACUCUGAUUCGGUGUCAUGUGCAUUGAAGCACGUGCUACACCCGAAAAAAAGGGCAGA---AGGCGCAAAA---------AGCGUACGAGAAAUGCAAAGUGCAGCCAG .(.((((..(((((((((((((((......)))))))..))).)))))....)))))...---.((((((...---------.((((.......))))....))).))).. ( -32.70, z-score = -2.41, R) >droSec1.super_0 988205 97 - 21120651 CGACUCUGAUUCGGUGUCAUGUGCAUUGAAGCACGUGCUACACCCG-AAAAAAGGGCAGA---AGGCGAAAAA---------AUCGUACGAGGAAUGCAAAGUGCAGCCA- .(.((((..(((((((((((((((......)))))))..))).)))-))...)))))...---.(((((....---------.))((((...(....)...)))).))).- ( -30.40, z-score = -1.94, R) >droYak2.chr3L 20997609 99 + 24197627 AGAUUCUGAUUCGGUGUCAUGUGCAUUGGAGCACGUGCUACACCCGAAAAAAAGAGCAGG---CGGCGCAAAA---------AGUGUUCGAGAAAUGCAAAGUGCAGCCAG .(.((((..(((((((((((((((......)))))))..))).)))))....))))).((---(.((((....---------.(((((.....)))))...)))).))).. ( -30.80, z-score = -1.47, R) >droEre2.scaffold_4784 20429719 97 + 25762168 UGAUUCUGAUUCGGUGGCAUGUGCAUUGAAGCACGUGCUACACCCG-AAAAAAGAGCAGG---AGGCGCAAAA---------AGUGUACGA-AAAUGCAAAGUGCAGCCAG ((.((((..(((((((((((((((......))))))))))...)))-))...))))))..---.((((((...---------..((((...-...))))...))).))).. ( -34.60, z-score = -3.45, R) >droAna3.scaffold_13337 15364385 111 + 23293914 CGAACCAGAUUUGGUGUCAUGUGCAUUGAAGCACGUGCUACACCCGAAAAGAACACAAGACCCCAACAAAAAAAAGUGUAAGAUUGUACGAAAAAUGCAAAGUGCAGCCAG ............((((((((((((......)))))))..)))))........((((...................))))..(..(((((............)))))..).. ( -22.11, z-score = -1.15, R) >dp4.chrXR_group8 329677 90 + 9212921 -AGUUGCGAUUCGGUGUCAUGUGCAUUGAAGCACGUGCUACACCCAAAAAAAAGACACGC-AGAACCCAAAAGAACUGCAAAGUGCAGCCAG------------------- -.((((((....((((((((((((......)))))))..)))))..............((-((............))))....))))))...------------------- ( -27.20, z-score = -2.74, R) >droWil1.scaffold_180698 3267992 96 + 11422946 AGUUCGCGAUUCAGUGUCAUGUGCAUUGAAGCACGUGCUAUACGAGCAGCAUAAGGAACACACUGUGACCCAA--GUGCAGACGCCAGCCAGUCAUCC------------- .(((.(((.((((((((.....))))))))(((((((((........)))))..((..(((...)))..))..--))))...))).))).........------------- ( -26.50, z-score = -0.54, R) >droMoj3.scaffold_6680 4690968 80 - 24764193 CUGCUGUGAUUCGGUGUCAUGUGCAUUGGAGCACGUGCUACACACAUAUACAAAAGACAA---GAAGAACAAA---------UUUAAGAGAG------------------- ....(((..(((..((((..((((......))))(((.....)))..........)))).---)))..)))..---------..........------------------- ( -17.60, z-score = -0.86, R) >droGri2.scaffold_15110 18193444 79 + 24565398 CAGUCGUGAUUCGGUGUCAUGUGCAUUGGAGCACGUGCUACAC-UAAAAAAAAAGUAAAA---AAAAAUCAAA---------GUACAGCCAG------------------- ..((..(((((..(((.(((((((......))))))).)))((-(........)))....---...)))))..---------..))......------------------- ( -14.80, z-score = -1.05, R) >consensus CGAUUCUGAUUCGGUGUCAUGUGCAUUGAAGCACGUGCUACACCCGAAAAAAAGAGAAGA___AGGCGCAAAA_________AGCGUACGAGAAAUGCAAAGUGCAGCCA_ .............(((((((((((......)))))))..)))).................................................................... (-12.40 = -12.41 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:12:17 2011