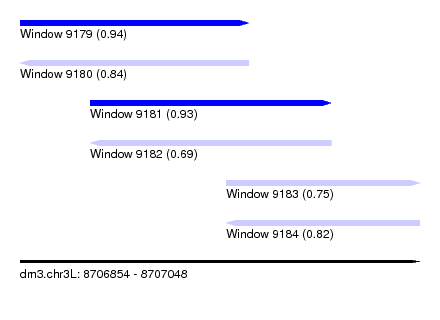
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,706,854 – 8,707,048 |
| Length | 194 |
| Max. P | 0.938130 |

| Location | 8,706,854 – 8,706,965 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 97.86 |
| Shannon entropy | 0.03800 |
| G+C content | 0.52859 |
| Mean single sequence MFE | -33.96 |
| Consensus MFE | -31.30 |
| Energy contribution | -31.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
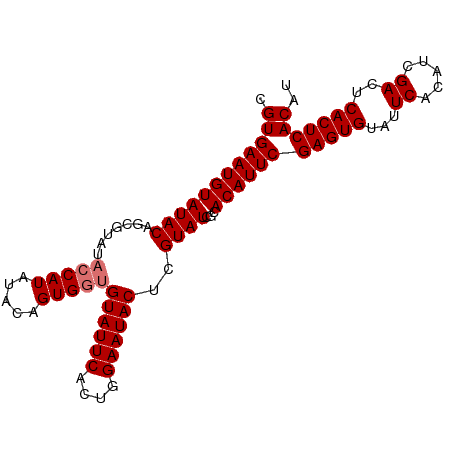
>dm3.chr3L 8706854 111 + 24543557 ACACCCCCUG---GAACCACCCGGCACACACGACUAAUUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUGUGAGUGAG .......(((---(......))))(((.((((.....((((((....))))))(((((((.....)))))))...(((((..(((.....))).))))).....)))).))).. ( -34.80, z-score = -2.56, R) >droEre2.scaffold_4784 20426037 111 - 25762168 ACACCCCCUC---GAACCACCCGGCACACACGACUAAUUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUGUGAGUGAG ..........---...((....))(((.((((.....((((((....))))))(((((((.....)))))))...(((((..(((.....))).))))).....)))).))).. ( -31.30, z-score = -1.96, R) >droYak2.chr3L 20993819 114 - 24197627 ACACCCCCCGCGCGAACCACCCGGCACACACGACUAAUUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUGUGAGUGAG .(((....((((((.......((((............((((((....))))))(((((((.....)))))))....))))...))))))((((..((....))..))))))).. ( -34.10, z-score = -1.93, R) >droSec1.super_0 984541 111 + 21120651 ACACCCCCUG---GAGCCACCCGGCACACACGACUAAUUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUGUGAGUGAG .......(((---(......))))(((.((((.....((((((....))))))(((((((.....)))))))...(((((..(((.....))).))))).....)))).))).. ( -34.80, z-score = -2.13, R) >droSim1.chr3L 8149622 111 + 22553184 ACACCCCCUG---GAACCACCCGGCACACACGACUAAUUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUGUGAGUGAG .......(((---(......))))(((.((((.....((((((....))))))(((((((.....)))))))...(((((..(((.....))).))))).....)))).))).. ( -34.80, z-score = -2.56, R) >consensus ACACCCCCUG___GAACCACCCGGCACACACGACUAAUUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUGUGAGUGAG .(((..................((((((...((....((((((....)))))))).))))))...((((((....(((((..(((.....))).)))))...)))))).))).. (-31.30 = -31.30 + 0.00)

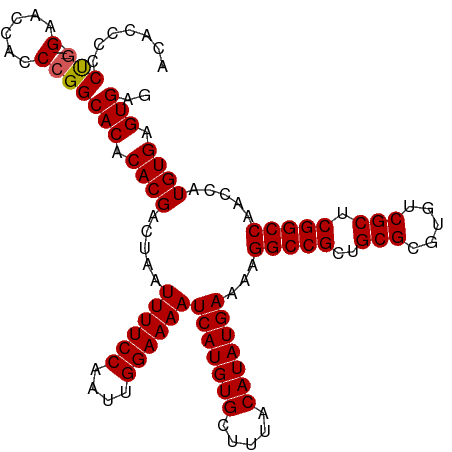
| Location | 8,706,854 – 8,706,965 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 97.86 |
| Shannon entropy | 0.03800 |
| G+C content | 0.52859 |
| Mean single sequence MFE | -40.66 |
| Consensus MFE | -38.02 |
| Energy contribution | -37.86 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.839243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8706854 111 - 24543557 CUCACUCACAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAAUUAGUCGUGUGUGCCGGGUGGUUC---CAGGGGGUGU ..((((((((((...(((((.(((.....)))..)))))......)))))...((((((((((((((....)))))....))))))))).(((((....))---).))))))). ( -40.00, z-score = -1.73, R) >droEre2.scaffold_4784 20426037 111 + 25762168 CUCACUCACAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAAUUAGUCGUGUGUGCCGGGUGGUUC---GAGGGGGUGU .(((((((((((...(((((.(((.....)))..)))))......)))))...((((((((((((((....)))))....)))))))))...))))))...---.......... ( -38.00, z-score = -1.22, R) >droYak2.chr3L 20993819 114 + 24197627 CUCACUCACAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAAUUAGUCGUGUGUGCCGGGUGGUUCGCGCGGGGGGUGU ..((((((((((...(((((.(((.....)))..)))))......)))))...((((((((((((((....)))))....))))))))).(((.(((...))).))).))))). ( -43.40, z-score = -1.86, R) >droSec1.super_0 984541 111 - 21120651 CUCACUCACAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAAUUAGUCGUGUGUGCCGGGUGGCUC---CAGGGGGUGU ..(((((...(((..(((((..((.(((((((.(((..((((........))))..).(((((((((....))))))))))).))))))).))..))))))---))..))))). ( -41.90, z-score = -2.03, R) >droSim1.chr3L 8149622 111 - 22553184 CUCACUCACAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAAUUAGUCGUGUGUGCCGGGUGGUUC---CAGGGGGUGU ..((((((((((...(((((.(((.....)))..)))))......)))))...((((((((((((((....)))))....))))))))).(((((....))---).))))))). ( -40.00, z-score = -1.73, R) >consensus CUCACUCACAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAAUUAGUCGUGUGUGCCGGGUGGUUC___CAGGGGGUGU ..((((((((((...(((((.(((.....)))..)))))......)))))...((((((((((((((....)))))....)))))))))(((....))).........))))). (-38.02 = -37.86 + -0.16)

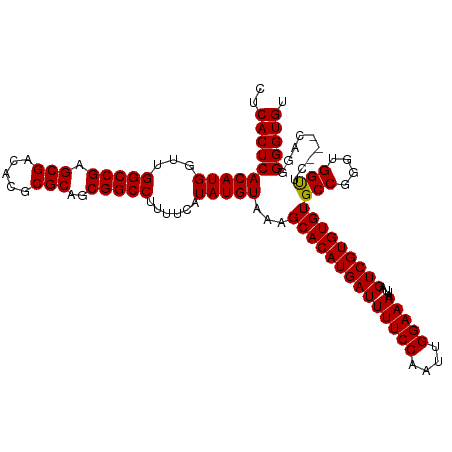
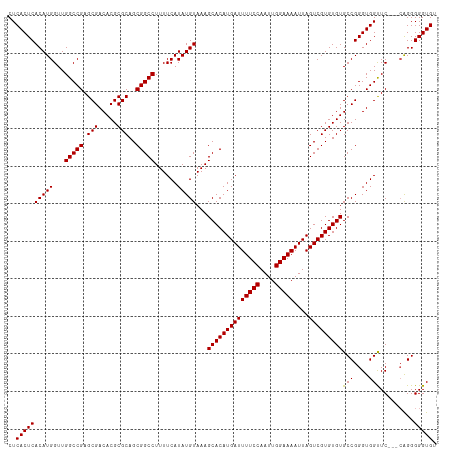
| Location | 8,706,888 – 8,707,005 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 94.87 |
| Shannon entropy | 0.10000 |
| G+C content | 0.45694 |
| Mean single sequence MFE | -35.63 |
| Consensus MFE | -32.63 |
| Energy contribution | -32.55 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.927855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
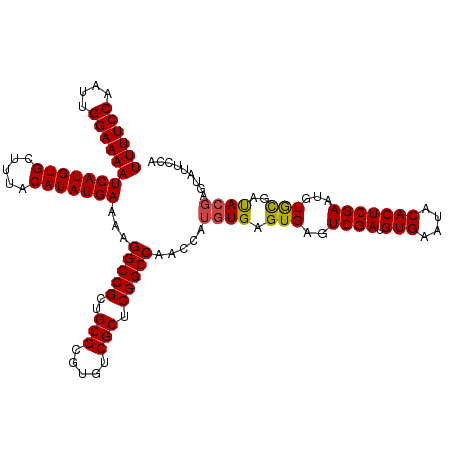
>dm3.chr3L 8706888 117 + 24543557 UUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUGUGAGUGAGUCGAUGUGAAUACACUCGAAUGUGCGAUACGAGUAUUCCA ((((((....))))))(((((((.....)))))))...((....(((.((((((((.(((....))...((((((..((.....))...))))))...).)))))))).)))..)). ( -36.10, z-score = -1.77, R) >droAna3.scaffold_13337 15361067 106 - 23293914 UUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUAUGAGUGAGUCGAUGUGAAUACACUCGAAUAUAUUCCA----------- ((((((....))))))(((((((.....)))))))...(((((..(((.....))).)))))........(((((..((((.(((....)))))))...)))))..----------- ( -33.30, z-score = -2.93, R) >droEre2.scaffold_4784 20426071 117 - 25762168 UUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUGUGAGUGAGUCGAUGUGAAUACACUCGAAUGUGCGAUACGAGUAUUCCA ((((((....))))))(((((((.....)))))))...((....(((.((((((((.(((....))...((((((..((.....))...))))))...).)))))))).)))..)). ( -36.10, z-score = -1.77, R) >droYak2.chr3L 20993856 117 - 24197627 UUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUGUGAGUGAGUCGAUGUGAAUACACUCGAAUGUGCGAUACGAGUAUUCCA ((((((....))))))(((((((.....)))))))...((....(((.((((((((.(((....))...((((((..((.....))...))))))...).)))))))).)))..)). ( -36.10, z-score = -1.77, R) >droSec1.super_0 984575 117 + 21120651 UUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUGUGAGUGAGUCGAUGUGAAUACACUCGAAUGUGCGAUACGAGUAUUCCA ((((((....))))))(((((((.....)))))))...((....(((.((((((((.(((....))...((((((..((.....))...))))))...).)))))))).)))..)). ( -36.10, z-score = -1.77, R) >droSim1.chr3L 8149656 117 + 22553184 UUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUGUGAGUGAGUCGAUGUGAAUACACUCGAAUGUGCGAUACGAGUAUUCCA ((((((....))))))(((((((.....)))))))...((....(((.((((((((.(((....))...((((((..((.....))...))))))...).)))))))).)))..)). ( -36.10, z-score = -1.77, R) >consensus UUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUGUGAGUGAGUCGAUGUGAAUACACUCGAAUGUGCGAUACGAGUAUUCCA ((((((....))))))(((((((.....)))))))...(((((..(((.....))).))))).....((((.(((..((((.(((....)))))))...)))..))))......... (-32.63 = -32.55 + -0.08)

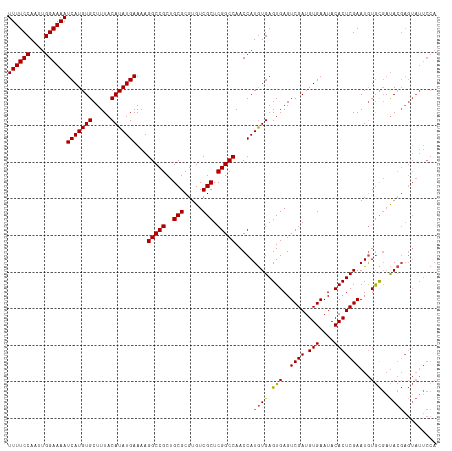
| Location | 8,706,888 – 8,707,005 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 94.87 |
| Shannon entropy | 0.10000 |
| G+C content | 0.45694 |
| Mean single sequence MFE | -31.58 |
| Consensus MFE | -28.38 |
| Energy contribution | -28.27 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.687800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8706888 117 - 24543557 UGGAAUACUCGUAUCGCACAUUCGAGUGUAUUCACAUCGACUCACUCACAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAA (((((...((((...((......(((((...((.....))..)))))(((((...(((((.(((.....)))..)))))......)))))...))..))))..)))))......... ( -31.60, z-score = -1.35, R) >droAna3.scaffold_13337 15361067 106 + 23293914 -----------UGGAAUAUAUUCGAGUGUAUUCACAUCGACUCACUCAUAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAA -----------..((((((((....)))))))).(((...((....(((((((..(((((.(((.....)))..)))))...)))))))...))...))).((((((....)))))) ( -31.50, z-score = -2.40, R) >droEre2.scaffold_4784 20426071 117 + 25762168 UGGAAUACUCGUAUCGCACAUUCGAGUGUAUUCACAUCGACUCACUCACAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAA (((((...((((...((......(((((...((.....))..)))))(((((...(((((.(((.....)))..)))))......)))))...))..))))..)))))......... ( -31.60, z-score = -1.35, R) >droYak2.chr3L 20993856 117 + 24197627 UGGAAUACUCGUAUCGCACAUUCGAGUGUAUUCACAUCGACUCACUCACAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAA (((((...((((...((......(((((...((.....))..)))))(((((...(((((.(((.....)))..)))))......)))))...))..))))..)))))......... ( -31.60, z-score = -1.35, R) >droSec1.super_0 984575 117 - 21120651 UGGAAUACUCGUAUCGCACAUUCGAGUGUAUUCACAUCGACUCACUCACAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAA (((((...((((...((......(((((...((.....))..)))))(((((...(((((.(((.....)))..)))))......)))))...))..))))..)))))......... ( -31.60, z-score = -1.35, R) >droSim1.chr3L 8149656 117 - 22553184 UGGAAUACUCGUAUCGCACAUUCGAGUGUAUUCACAUCGACUCACUCACAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAA (((((...((((...((......(((((...((.....))..)))))(((((...(((((.(((.....)))..)))))......)))))...))..))))..)))))......... ( -31.60, z-score = -1.35, R) >consensus UGGAAUACUCGUAUCGCACAUUCGAGUGUAUUCACAUCGACUCACUCACAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAA ...............((......(((((...((.....))..)))))(((((...(((((.(((.....)))..)))))......)))))...))......((((((....)))))) (-28.38 = -28.27 + -0.11)
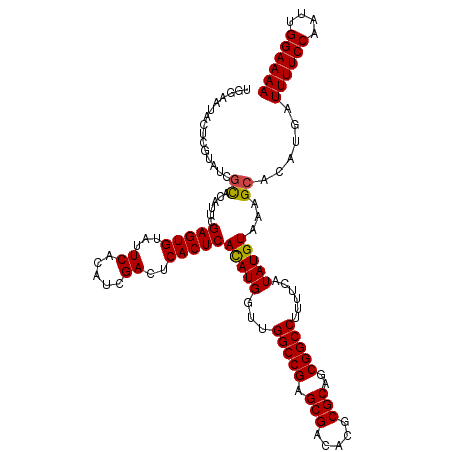
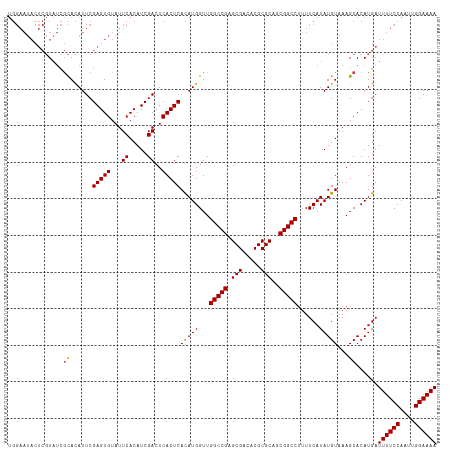
| Location | 8,706,954 – 8,707,048 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 94.86 |
| Shannon entropy | 0.08500 |
| G+C content | 0.41145 |
| Mean single sequence MFE | -28.35 |
| Consensus MFE | -24.21 |
| Energy contribution | -24.52 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.748385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8706954 94 + 24543557 AUGUGAGUGAGUCGAUGUGAAUACACUCGAAUGUGCGAUACGAGUAUUCCAGUGAAUACACCACUGUAUAUGGUAUAUGCUGUAUACAUUCACG ..(((((((..((((.(((....)))))))....(((((((..((((..(((((.......))))).)))).)))).)))......))))))). ( -27.20, z-score = -1.30, R) >droEre2.scaffold_4784 20426137 85 - 25762168 AUGUGAGUGAGUCGAUGUGAAUACACUCGAAUGUGCGAUACGAGUAUUCCAGUGAAUAC--CACUGUAUAUG-------CUGUAUACAUUCACG ..(((((((..((((.(((....))))))).......((((.(((((..(((((.....--)))))...)))-------)))))).))))))). ( -27.60, z-score = -2.77, R) >droSec1.super_0 984641 94 + 21120651 AUGUGAGUGAGUCGAUGUGAAUACACUCGAAUGUGCGAUACGAGUAUUCCAGUGAAUACACCACUGUAUAUGGUAUACGCUGUAUACAUUCACG ..(((((((..((((.(((....)))))))....(((((((..((((..(((((.......))))).)))).)))).)))......))))))). ( -29.30, z-score = -1.90, R) >droSim1.chr3L 8149722 94 + 22553184 AUGUGAGUGAGUCGAUGUGAAUACACUCGAAUGUGCGAUACGAGUAUUCCAGUGAAUACACCACUGUAUAUGGUAUACGCUGUAUACAUUCACG ..(((((((..((((.(((....)))))))....(((((((..((((..(((((.......))))).)))).)))).)))......))))))). ( -29.30, z-score = -1.90, R) >consensus AUGUGAGUGAGUCGAUGUGAAUACACUCGAAUGUGCGAUACGAGUAUUCCAGUGAAUACACCACUGUAUAUGGUAUACGCUGUAUACAUUCACG ..(((((((..((((.(((....))))))).............((((((....))))))..)))((((((((((....)))))))))).)))). (-24.21 = -24.52 + 0.31)

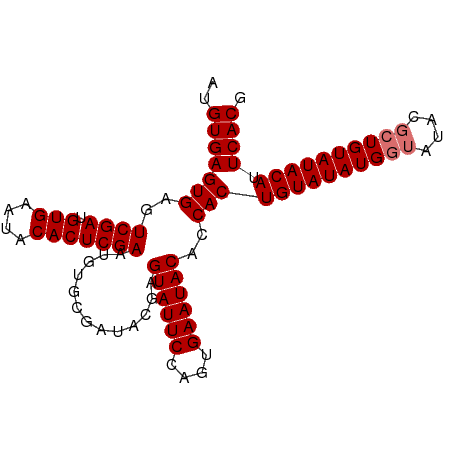
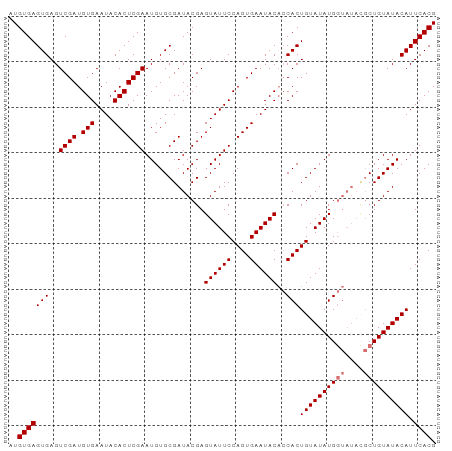
| Location | 8,706,954 – 8,707,048 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 94.86 |
| Shannon entropy | 0.08500 |
| G+C content | 0.41145 |
| Mean single sequence MFE | -23.95 |
| Consensus MFE | -22.58 |
| Energy contribution | -22.70 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.820222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8706954 94 - 24543557 CGUGAAUGUAUACAGCAUAUACCAUAUACAGUGGUGUAUUCACUGGAAUACUCGUAUCGCACAUUCGAGUGUAUUCACAUCGACUCACUCACAU .((((((.....(((.(((((((((.....)))))))))...)))..(((((((...........)))))))))))))................ ( -24.70, z-score = -1.78, R) >droEre2.scaffold_4784 20426137 85 + 25762168 CGUGAAUGUAUACAG-------CAUAUACAGUG--GUAUUCACUGGAAUACUCGUAUCGCACAUUCGAGUGUAUUCACAUCGACUCACUCACAU .((((.((((((...-------..))))))(((--((((((....)))))).............(((((((....))).))))..))))))).. ( -20.90, z-score = -1.42, R) >droSec1.super_0 984641 94 - 21120651 CGUGAAUGUAUACAGCGUAUACCAUAUACAGUGGUGUAUUCACUGGAAUACUCGUAUCGCACAUUCGAGUGUAUUCACAUCGACUCACUCACAU .((((.((......(((.((((..(((.((((((.....))))))..)))...)))))))....(((((((....))).))))..)).)))).. ( -25.10, z-score = -1.48, R) >droSim1.chr3L 8149722 94 - 22553184 CGUGAAUGUAUACAGCGUAUACCAUAUACAGUGGUGUAUUCACUGGAAUACUCGUAUCGCACAUUCGAGUGUAUUCACAUCGACUCACUCACAU .((((.((......(((.((((..(((.((((((.....))))))..)))...)))))))....(((((((....))).))))..)).)))).. ( -25.10, z-score = -1.48, R) >consensus CGUGAAUGUAUACAGCGUAUACCAUAUACAGUGGUGUAUUCACUGGAAUACUCGUAUCGCACAUUCGAGUGUAUUCACAUCGACUCACUCACAU .((((((((((((.......(((((.....)))))((((((....))))))..))))...))))))(((((...((.....))..))))))).. (-22.58 = -22.70 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:12:16 2011