| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,640,047 – 8,640,140 |
| Length | 93 |
| Max. P | 0.722017 |

| Location | 8,640,047 – 8,640,140 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 62.33 |
| Shannon entropy | 0.80673 |
| G+C content | 0.52590 |
| Mean single sequence MFE | -29.13 |
| Consensus MFE | -10.14 |
| Energy contribution | -10.53 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.72 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.722017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
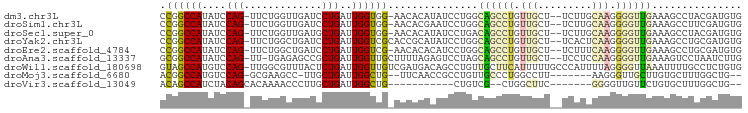
>dm3.chr3L 8640047 93 + 24543557 CCGGCCAUAUCCAG-UUCUGGUUGAUCCUGAUUGGUGG-AACACAUAUCCUGGCAGCCUGUUGCU--UCUUGCAAGGGGUUGAAAGCCUACGAUGUG ....(((...((((-((..((.....)).)))))))))-....((((((..((((((((.((((.--....)))).)))))....)))...)))))) ( -30.00, z-score = -0.95, R) >droSim1.chr3L 8080023 93 + 22553184 CCGGCCAUAUCCAG-UUCUGGUUGAUCCUGAUUGGUGG-AACACGAAUCCUGGCAGCCUGUUGCU--UCUUGCAAGGGGUUGAAAGCCUUCGAUGUG ....(((...((((-((..((.....)).)))))))))-.(((((((..((..((((((.((((.--....)))).))))))..))..)))).))). ( -32.30, z-score = -1.60, R) >droSec1.super_0 918327 93 + 21120651 CCGGCCAUAUCCAG-UUCUGGUUGAUGCUGAUUGGUGG-AACACAUAUCCUGACAGCCUGUUGCU--UCUUGCAAGGGGUUGAAAGCCUACGAUGUG .(.((((....(((-(((.....)).))))..)))).)-....((((((((..((((((.((((.--....)))).))))))..)).....)))))) ( -28.90, z-score = -0.98, R) >droYak2.chr3L 20925452 94 - 24197627 CCGGCCAUAUCCAG-UUCUGGCUGAUCCUGAUUGGUCGCACCGCAUAUCCUGGCAGCCUGUUGCU--UCACUCAAGGGGUUGAAAGCCUGCGAUGUG ..(((((.((((((-(....)))).....))))))))(((.((((....((..((((((.(((..--.....))).))))))..))..)))).))). ( -28.90, z-score = 0.12, R) >droEre2.scaffold_4784 20356874 93 - 25762168 CCGGCCAUAUCCAG-UUCUGGCUGAUCCUGAUUGGUCG-AACACACAUCCUGGCAGCCUGUUGCU--UCUUUCAAGGGGUUGAAAGCCUGCGAUGUG .((((((.((((((-(....)))).....)))))))))-....(((((((.((((((((.(((..--.....))).)))))....))).).)))))) ( -30.10, z-score = -0.92, R) >droAna3.scaffold_13337 15297547 93 - 23293914 GCGGCCAUAUCCAG-UU-UGAGAGCCGCUGAUUGGUUGCUUUUAGAGUCCUAGCAGCCUGUUGCU--UCCUCCAAGGGGUUGAAAGUCCUAAUCUUG (((((....((...-..-.))..))))).((((((..((((((((..(((((((((....)))))--.......)))).)))))))).))))))... ( -27.01, z-score = -0.58, R) >droWil1.scaffold_180698 3167124 96 - 11422946 GUAGCCAUGUCCAG-UUGGCGUUUACUCUGAUUGGUUGUCGAUGACAGCCUGUUGCUUCAUUUUUGCCCAUUUUAGGGGUUAAAUUUUGCCUCUGUG (((((((.......-.)))).........(((.(((((((...))))))).)))..........)))......(((((((........))))))).. ( -23.80, z-score = -0.82, R) >droMoj3.scaffold_6680 4606259 84 + 24764193 ACGGCCAUGUCCAG-GCGAAGCC-UUGCUGAUUGGCUG--UUCAACCGCCUGUUGCCCUGGCCUU-------AAGGGUUGCUUGUGCUUUGGCUG-- .((((((.(..(((-((((..((-(((......(((((--..((((.....))))..).)))).)-------)))).)))))))..)..))))))-- ( -31.40, z-score = -1.05, R) >droVir3.scaffold_13049 6292391 75 + 25233164 ACAGCCAUCUACAGCACAAAACCCUUGCUGAUUGGCUG-----------CUGUCG--CUGGCUUC-------GGGGUUGUUCUGUGCUUUGGCUG-- .((((((.....((((((.((((((.(((...((((..-----------..))))--..)))...-------))))))....)))))).))))))-- ( -29.80, z-score = -2.79, R) >consensus CCGGCCAUAUCCAG_UUCUGGCUGAUCCUGAUUGGUUG_AACACACAUCCUGGCAGCCUGUUGCU__UCUUGCAAGGGGUUGAAAGCCUUCGAUGUG .((((((....(((.............)))..))))))...............((((((.(((.........))).))))))............... (-10.14 = -10.53 + 0.39)

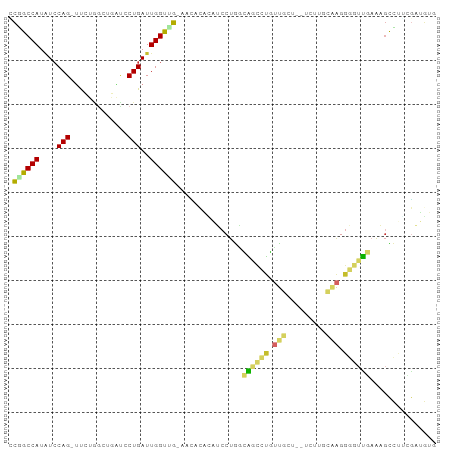
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:12:04 2011