| Sequence ID | dm3.chr3L |
|---|---|
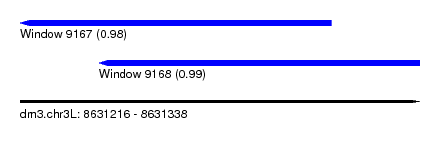
| Location | 8,631,216 – 8,631,338 |
| Length | 122 |
| Max. P | 0.986132 |

| Location | 8,631,216 – 8,631,311 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 68.35 |
| Shannon entropy | 0.63135 |
| G+C content | 0.43745 |
| Mean single sequence MFE | -23.77 |
| Consensus MFE | -12.77 |
| Energy contribution | -13.43 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.977672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
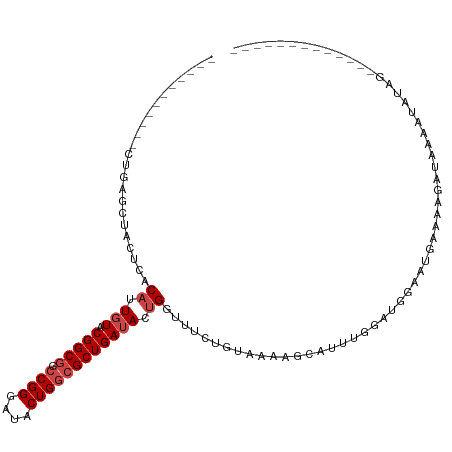
>dm3.chr3L 8631216 95 - 24543557 --------AAUCCGCGCUACUCACAUUGUACGGCGGCCGGGAUACUGGCGCUGAUACUGGUUUCUGUAAUAGCAUUUGGAUGGAGUGAACAGAUAAGAUAUAG------------- --------.(((.(..(.((((.((((((((((((.((((....))))))))).)))..(((........))).....)))))))))..).))).........------------- ( -24.30, z-score = -0.31, R) >droSim1.chr3L 8071227 93 - 22553184 ----------GCCGAACUACUCACAUUGUACGGCGGCCGGGAUACUGGCGCUGAUACUGGUUUCUGUAAUAGCAUUUGGAUGGAGUGAACAGAUAAGAUAUAG------------- ----------((((.((..........)).)))).(((((....)))))((((.(((........))).))))(((((...........))))).........------------- ( -24.20, z-score = -0.96, R) >droSec1.super_0 909700 95 - 21120651 --------AAGCCGGGCUACUCACAUUGUACGGCGGCCGGGAUACUGGCGCUGAUACUGGUUUCUGUAAUAGCAUUUGGAUGGAGUGAACAGAUAAGAUAUAG------------- --------.((((((..(((.......)))(((((.((((....)))))))))...))))))(((((.....(((((.....))))).)))))..........------------- ( -26.40, z-score = -0.99, R) >droYak2.chr3L 20916413 97 + 24197627 ------UAAGUGUGAGCUACUUACAUUGUACGGCGGCCGGGAUACUGGCGCUGAUACUGGUUUCUGUAAUAGCAUUUGGAUGGAGUGAACAGAUAAGAUACAG------------- ------..((((((((...))))))))((((((((.((((....)))))))))...((....(((((.....(((((.....))))).)))))..)).)))..------------- ( -26.90, z-score = -1.60, R) >droEre2.scaffold_4784 20346852 94 + 25762168 ---------AGCUGAGCUACUCACAUUGUACGGCGGCCGGGAUACUGGCGCUGAUACUGGUUUCUGUAAUAGCAUUUGGAUGAAGUAAACAGAAAUAACUCUG------------- ---------....(((.......((.(((.(((((.((((....)))))))))))).))((((((((.....(((....)))......))))))))..)))..------------- ( -25.70, z-score = -1.41, R) >droAna3.scaffold_13337 15289064 92 + 23293914 ------------GGAGCUACUCACAUUGUACGGCGGCCGGGAUACUGGCGCUGAUACUGGUUUCUGUAAGAUCAUUCAGUGGGGAAUA-AAUAAAUAAUAUUCAA----------- ------------.......(((((...((((((((.((((....))))))))).)))((((((.....))))))....)))))(((((-.........)))))..----------- ( -25.60, z-score = -2.20, R) >dp4.chrXR_group8 245294 101 + 9212921 ------------UAAACCACUCACAUUGUACGGCGGACGGGAUACUGGGGCUGAUACCGGUUUCUGUAAAGGCAUU---UCGGAAAUACAAGAUAUAUUAUUGAGAUAGGAAAUAC ------------....((.............(.(..((((((.(((((........)))))))))))...).)(((---((((.((((.......)))).))))))).))...... ( -20.50, z-score = -0.86, R) >droPer1.super_50 237979 101 - 557249 ------------UAAACCACUCACAUUGUACGGCGGACGGGAUACUGGGGCUGAUACCGGUUUCUGUAAAUGCAUU---UGGGAAAUACAAGAUAUAUUAUUGAGAUAGGAAAUAC ------------....((.((((...((((..(((.((((((.(((((........)))))))))))...))).((---((.......)))).))))....))))...))...... ( -21.50, z-score = -1.33, R) >droWil1.scaffold_180698 3153971 90 + 11422946 GAAAGAAGUUGAACAAGUACUUACAUUGUACGGCGGCCGGGAUACUGGCGCUGAUACUGGUUUCUAAAGAAAAG-----AGAGAACGAGAAAAUG--------------------- ((((..(((.......((((.......))))((((.((((....))))))))...)))..))))..........-----................--------------------- ( -19.20, z-score = -1.53, R) >droVir3.scaffold_13049 6283790 95 - 25233164 -----------CUCGGUAACUUACAUUGUACGGCGGCCGGGAUACUGGCGCUGAAACUGGUUUCUGUAAAAG-GUAUAUGGGGCAUGUAAAAUACAAAGAAAGAACU--------- -----------((..(((..((((((....(((((.((((....)))))))))......((..(((((....-.)))).)..))))))))..)))..))........--------- ( -24.90, z-score = -1.97, R) >droMoj3.scaffold_6680 4597034 93 - 24764193 -----------UUCUGUUACUUACAUUGUACGGCGGCCGGGAUACUGGCGCUGAUACUGGCUUCUGUAAAGGCAUUUAUAAGGUAUGUAGAA-AUAAAGAUAACA----------- -----------.(((.....((((((.((((((((.((((....))))))))).)))..((((.((((((....))))))))))))))))..-....))).....----------- ( -23.40, z-score = -1.17, R) >droGri2.scaffold_15110 18112922 90 + 24565398 -----------CUCUGUUACUUACAUUGUACGGCGGCCGGGAUACUGGCGCUGAAAAUGGUUUCUGUAAAAGUACAUUUAAAUGAUGGAAAA-AAAAAAAUA-------------- -----------.(((((((.(((...(((((((((.((((....))))))))((((....)))).......)))))..))).)))))))...-.........-------------- ( -22.60, z-score = -3.13, R) >consensus ___________CUGAGCUACUCACAUUGUACGGCGGCCGGGAUACUGGCGCUGAUACUGGUUUCUGUAAAAGCAUUUGGAUGGAAUGAAAAGAUAAAAUAUAG_____________ .......................((.(((.(((((.((((....)))))))))))).))......................................................... (-12.77 = -13.43 + 0.67)

| Location | 8,631,240 – 8,631,338 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 66.68 |
| Shannon entropy | 0.72983 |
| G+C content | 0.43377 |
| Mean single sequence MFE | -23.73 |
| Consensus MFE | -12.77 |
| Energy contribution | -13.43 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.986132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
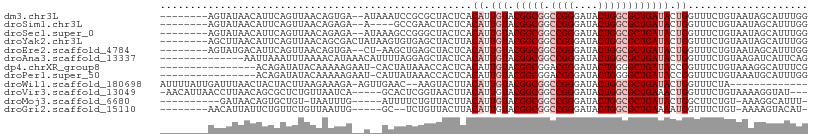
>dm3.chr3L 8631240 98 - 24543557 --------AGUAUAACAUUCAGUUAACAGUGA--AUAAAUCCGCGCUACUCACAUUGUACGGCGGCCGGGAUACUGGCGCUGAUACUGGUUUCUGUAAUAGCAUUUGG --------..(((.(((.(((((...((((..--.......(((..(((.......)))..)))(((((....)))))))))..)))))....))).)))........ ( -23.30, z-score = -0.01, R) >droSim1.chr3L 8071251 94 - 22553184 --------AGUAUAACAUUCAGUUAACAGAGA--A----GCCGAACUACUCACAUUGUACGGCGGCCGGGAUACUGGCGCUGAUACUGGUUUCUGUAAUAGCAUUUGG --------.............((((..(.(((--(----(((((.....)).....((((((((.((((....))))))))).))).))))))).)..))))...... ( -27.70, z-score = -2.02, R) >droSec1.super_0 909724 98 - 21120651 --------AGUAUAACAUUCAGUUAACAGAGA--AUAAAGCCGGGCUACUCACAUUGUACGGCGGCCGGGAUACUGGCGCUGAUACUGGUUUCUGUAAUAGCAUUUGG --------.............((((((((((.--.....(((((..(((.......)))(((((.((((....)))))))))...)))))))))))..))))...... ( -26.80, z-score = -1.10, R) >droYak2.chr3L 20916437 100 + 24197627 --------AGCUUAACAUUCAGUUAACAGCGACUAUAAGUGUGAGCUACUUACAUUGUACGGCGGCCGGGAUACUGGCGCUGAUACUGGUUUCUGUAAUAGCAUUUGG --------.(((((((.....))))((((.(((((..((((((((...))))))))((((((((.((((....))))))))).)))))))).))))...)))...... ( -32.50, z-score = -2.48, R) >droEre2.scaffold_4784 20346876 97 + 25762168 --------AGUAUGACAUUCAGUUAACAGUGA--CU-AAGCUGAGCUACUCACAUUGUACGGCGGCCGGGAUACUGGCGCUGAUACUGGUUUCUGUAAUAGCAUUUGG --------.............((((((((.((--((-(.(.((((...)))))...((((((((.((((....))))))))).)))))))).))))..))))...... ( -27.00, z-score = -0.82, R) >droAna3.scaffold_13337 15289089 94 + 23293914 --------------AAUUAAAUUUAAAACAUAAACAUUUUAGGAGCUACUCACAUUGUACGGCGGCCGGGAUACUGGCGCUGAUACUGGUUUCUGUAAGAUCAUUCAG --------------.........................(((((((((........((((((((.((((....))))))))).))))))))))))............. ( -21.60, z-score = -1.98, R) >dp4.chrXR_group8 245328 91 + 9212921 ----------------ACAGAUAUACAAAAAGAAU-CACUAUAAACCACUCACAUUGUACGGCGGACGGGAUACUGGGGCUGAUACCGGUUUCUGUAAAGGCAUUUCG ----------------.......(((((...((..-.............))...))))).(.(..((((((.(((((........)))))))))))...).)...... ( -14.76, z-score = 0.53, R) >droPer1.super_50 238013 91 - 557249 ----------------ACAGAUAUACAAAAAGAAU-CAUUAUAAACCACUCACAUUGUACGGCGGACGGGAUACUGGGGCUGAUACCGGUUUCUGUAAAUGCAUUUGG ----------------.(((((.(((((...((..-.............))...)))))..(((.((((((.(((((........)))))))))))...)))))))). ( -18.86, z-score = -0.98, R) >droWil1.scaffold_180698 3153995 92 + 11422946 AUUUUAUUGAUUUAACUACUACUUAAGAAAGA-AGUUGAAC--AAGUACUUACAUUGUACGGCGGCCGGGAUACUGGCGCUGAUACUGGUUUCUA------------- ......(((.(((((((.((.........)).-))))))))--))......((...((((((((.((((....))))))))).)))..)).....------------- ( -21.40, z-score = -2.00, R) >droVir3.scaffold_13049 6283820 99 - 25233164 -AACAUUAACCUUAACAGCGCUCUGUUAAUCA-----GCACUCGGUAACUUACAUUGUACGGCGGCCGGGAUACUGGCGCUGAAACUGGUUUCUGUAAAAGGUAU--- -.(((..(((((((((((....)))))))(((-----((.(.(.(((.(.......)))).).)(((((....))))))))))....))))..))).........--- ( -28.70, z-score = -1.98, R) >droMoj3.scaffold_6680 4597061 90 - 24764193 ----------GAUAACAGUGCUGU-UAAUUUG-----AUUUUCUGUUACUUACAUUGUACGGCGGCCGGGAUACUGGCGCUGAUACUGGCUUCUGU-AAAGGCAUUU- ----------..((((((....((-(.....)-----))...))))))....((.(((.(((((.((((....)))))))))))).))((((....-..))))....- ( -22.80, z-score = -1.12, R) >droGri2.scaffold_15110 18112946 91 + 24565398 --------AACAUUAUUCUGUUCUGUUAAUUG-----GC--UCUGUUACUUACAUUGUACGGCGGCCGGGAUACUGGCGCUGAAAAUGGUUUCUGU-AAAAGUACAU- --------......................((-----..--.((.((((...((((...(((((.((((....)))))))))..))))......))-)).))..)).- ( -19.30, z-score = -0.35, R) >consensus ________AGUAUAACAUUCAUUUAACAAAGA__A__AAUCUGAGCUACUCACAUUGUACGGCGGCCGGGAUACUGGCGCUGAUACUGGUUUCUGUAAAAGCAUUUGG ....................................................((.(((.(((((.((((....)))))))))))).)).................... (-12.77 = -13.43 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:12:03 2011