| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,582,222 – 8,582,328 |
| Length | 106 |
| Max. P | 0.866671 |

| Location | 8,582,222 – 8,582,328 |
|---|---|
| Length | 106 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 66.59 |
| Shannon entropy | 0.71835 |
| G+C content | 0.57547 |
| Mean single sequence MFE | -38.55 |
| Consensus MFE | -16.29 |
| Energy contribution | -16.59 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.866671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
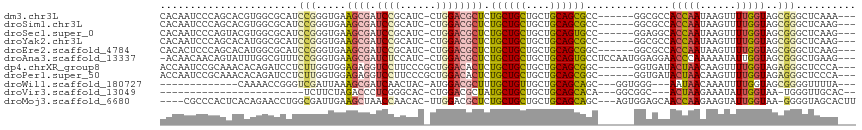
>dm3.chr3L 8582222 106 + 24543557 CACAAUCCCAGCACGUGGCGCAUCCGGGUGAAGCGAUCCGCAUC-CUGGACGCUCUGCUGCUGCUGCAGCGCC------GGCGCCACCAAUAAGUUUUGGUAGCGGGCUCAAA--- ......(((.((....(((((.((((((....(((...)))..)-))))).))...(((((....))))))))------.))((.(((((......))))).)))))......--- ( -43.40, z-score = -1.00, R) >droSim1.chr3L 8023544 106 + 22553184 CACAAUCCCAGCACGUGGCGCAUCCGGGUGAAGCGAUCCGCAUC-CUGGACGCUCUGCUGCUGCUGCAGCGCC------GGCGCCACCAAUAAGUUUUGGUAGCGGGCUCAAG--- ......(((.((....(((((.((((((....(((...)))..)-))))).))...(((((....))))))))------.))((.(((((......))))).)))))......--- ( -43.40, z-score = -1.02, R) >droSec1.super_0 861083 106 + 21120651 CACAAUCCCAGUACGUGGCGCAUCCGGGUGAAGCGAUCCGCAUC-CUGGACGCUCUGCUGCUGCUGCAGUGCC------GGAGGCACCAAUAAGUUUUGGUAGCGGGCUCAAG--- ......(((((((.(..(.((.((((((....(((...)))..)-))))).)).)..)))))(((((.(((((------...)))))(((......)))))))))))......--- ( -40.30, z-score = -0.62, R) >droYak2.chr3L 20866310 106 - 24197627 CACAAUCCCAGCACAUGGCGCAUCCGGGUGAAGCGAUCCGCAUC-CUGGACGCUCUGCUGCUGCUGCAGCGCC------GGCGCCACCAAUAAGUUUUGGUAGCGGGCUCAAG--- ......(((.((....(((((.((((((....(((...)))..)-))))).))...(((((....))))))))------.))((.(((((......))))).)))))......--- ( -43.40, z-score = -1.31, R) >droEre2.scaffold_4784 20297019 106 - 25762168 CACACUCCCAGCACAUGGCGCAUCCGGGUGAAGCGAUCCGCAUC-CUGGACGCUCUGCUGCUGCUGCAGCGGC------GGCGCCACCAAUAAGUUUUGGUAGCGGGCUCAAG--- ......(((.((....(((((.((((((....(((...)))..)-)))))((((..(((((....))))))))------))))))(((((......))))).)))))......--- ( -46.80, z-score = -1.97, R) >droAna3.scaffold_13337 15236383 111 - 23293914 -ACAACAACAGUAUUUGGCGUUUCCGGGUGAAGCGAUCUCCAUC-CUGGACGCUCUGCUGCUGCUGCAGUGCCUCCAAUGGAGGAACCCAAAAAUAUUGGUAGCGGGCUGAAG--- -.......((((((((((((((((.....)))))).(((((((.-.((((.((.((((.......)))).)).)))))))))))...)))..))))))).(((....)))...--- ( -36.00, z-score = -0.72, R) >dp4.chrXR_group8 190604 107 - 9212921 ACCAAUCCGCAAACACAGAUCCUCUUGGUGGAGAGGUCCUUCCCGCUGGACACUCUGCUGCUGCUGCAGCGGC------GGUGAUACUAACAAGUUUUGGUAGAGGGCUCCCA--- .................((((((((..((((.(((....)))))))..).(((((((((((....))))))).------)))).((((((......))))))))))).))...--- ( -35.30, z-score = -0.07, R) >droPer1.super_50 184268 107 + 557249 ACCAAUCCGCAAACACAGAUCCUCUUGGUGGAGAGGUCCUUCCCGCUGGACACUCUGCUGCUGCUGCAGCGGC------GGUGAUACUAACAAGUUUUGGUAGAGGGCUCCCA--- .................((((((((..((((.(((....)))))))..).(((((((((((....))))))).------)))).((((((......))))))))))).))...--- ( -35.30, z-score = -0.07, R) >droWil1.scaffold_180727 1476538 93 - 2741493 -------------CAAAACCGGGUCGAUUAAAGCGAUCAACUAC-AUGGACGCUUUGCUGUUGCUGCAGCAGC---GGUGGG---AAUAACAAAUUUUGGUAGCGGGGUUUUA--- -------------.((((((.(((((.......)))))......-.....(((((..(((((((....)))))---))..).---.....((.....))..)))).)))))).--- ( -26.70, z-score = -1.35, R) >droVir3.scaffold_13049 6238513 82 + 25233164 ------------------------UCUUCUAGACCCUCGGGCAC-CUGGACGCUAUGCUGCUGCUGCAGCACA---GGCGGC---ACUAAGAAAUAUUGGUAA-UGGGUUGCAC-- ------------------------.......(((((((((....-)))).((((.((((((....))))))..---))))..---(((((......)))))..-.)))))....-- ( -27.50, z-score = -1.02, R) >droMoj3.scaffold_6680 4553933 107 + 24764193 ----CGCCCACUCACAGAACCUGGCGAUUGAAGCUAACCAACAC-UUGGACGCUCUGCUGCUGCUGCAGCAGC---AGUGGAGCAACCAAGAAGUAUUGGUAA-GGGGUAGCACUU ----.((((............((((.......))))(((((..(-((((..((((..(((((((....)))))---))..))))..))))).....)))))..-.))))....... ( -45.90, z-score = -3.59, R) >consensus CACAAUCCCAGCACAUGGCGCAUCCGGGUGAAGCGAUCCGCAUC_CUGGACGCUCUGCUGCUGCUGCAGCGCC______GGCGCCACCAAUAAGUUUUGGUAGCGGGCUCAAA___ .......................(((.....((((.((((......)))))))).((((((....))))))..............(((((......)))))..))).......... (-16.29 = -16.59 + 0.30)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:11:55 2011