| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,574,340 – 8,574,436 |
| Length | 96 |
| Max. P | 0.908556 |

| Location | 8,574,340 – 8,574,436 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 69.88 |
| Shannon entropy | 0.58505 |
| G+C content | 0.37692 |
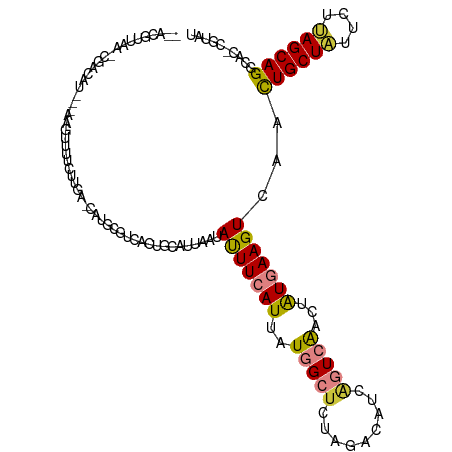
| Mean single sequence MFE | -24.57 |
| Consensus MFE | -11.19 |
| Energy contribution | -11.30 |
| Covariance contribution | 0.11 |
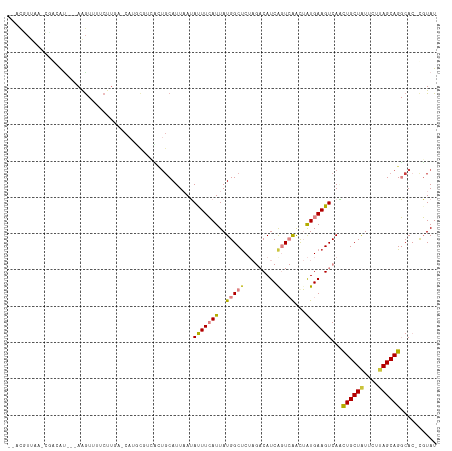
| Combinations/Pair | 1.33 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
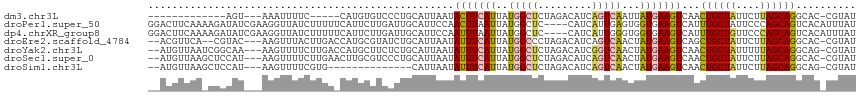
>dm3.chr3L 8574340 96 + 24543557 -------------AGU---AAAUUUUC-----CAUGUGUCCCUGCAUUAAUACUUCAUUAUGGCUCUAGACAUCAGUCAAUUAUGAAGUCAACUGCUAUUCUUAGCAGGCAC-CGUAU -------------...---........-----...((((............(((((((..(((((.........)))))...)))))))...((((((....))))))))))-..... ( -20.80, z-score = -1.92, R) >droPer1.super_50 175668 114 + 557249 GGACUUCAAAAGAUAUCGAAGGUUAUCUUUUUCAUUCUUGAUUGCAUUCCAACUUAAUUAUGGCUC----CAUCAUUGAGUGGUGAAGUCAUUUGCUAUUCCCAGCAGUCACAUUUAU .((((((((((((((.(....).)))))))..................(((.((((((.(((....----))).))))))))))))))))..(((((......))))).......... ( -28.90, z-score = -2.75, R) >dp4.chrXR_group8 182048 114 - 9212921 GGACUUCAAAAGAUAUCGAAGGUUAUCUUUUUCAUUCUUGAUUGCAUUCCAAUUUAAUUAUGGCUC----CAUCAUUGGGUGGUGAAGUCAUUUGCUGUUCCCAGCAGUCACAUUUAU .((((((((((((((.(....).))))))).((((....(((((.....))))).....))))..(----((((....))))))))))))..((((((....)))))).......... ( -28.70, z-score = -2.02, R) >droEre2.scaffold_4784 20289163 110 - 25762168 --ACGUUCA--CGUAC---AAGUUUACUUGACCAUGCGUAUCUGCAUUAAUAUUUCAUUAUGGCCCUAGACAUCAGUCAACUAUGAAGUCAGCUGCUAUUCUUAGCAGGCAC-CGUAU --((((...--.((.(---(((....))))))...))))...................(((((.....(((.(((........))).)))..((((((....))))))...)-)))). ( -24.20, z-score = -1.18, R) >droYak2.chr3L 20858364 112 - 24197627 --AUGUUAAUCGGCAA---AAGUUUUCUUGACCAUGCUUCUCUGCAUUAAUAUUUCAUUAUGGCUCUAGACAUCGGUCAACUAUGAAGUCAACUGCUAUUUUUAGCAGGCAG-CGUAU --.........((((.---..(((.....)))..)))).(.((((......(((((((..(((((.........)))))...)))))))...((((((....))))))))))-.)... ( -24.10, z-score = -0.89, R) >droSec1.super_0 853431 112 + 21120651 --AUGUUAAGCUCCAU---AAGUUUUCUUGAACUUGCGUCCCUGCAUUAAUAUUUCAUUAUGGCUCUAGACAUCAGUCAACUAUGAAGUCAACUGCUAUUCUUAGCAGGCAC-CGUAU --(((((((((..(.(---((((((....))))))).).....)).))))))).....(((((.....(((.(((........))).)))..((((((....))))))...)-)))). ( -25.10, z-score = -1.78, R) >droSim1.chr3L 8015855 98 + 22553184 --AUGUUAAGCUCCAU---AAGUUUUCGUG--------------CAUUAAUAUUUCAUUAUGGCUCUAGACAUCAGUCAACUAUGAAGUCAACUGCUAUUCUUAGCAGGCAG-CGUAU --((((((((((....---.)))))....(--------------(......(((((((..(((((.........)))))...)))))))...((((((....))))))))))-))).. ( -20.20, z-score = -0.81, R) >consensus __ACGUUAA_CGACAU___AAGUUUUCUUGA_CAUGCGUCACUGCAUUAAUAUUUCAUUAUGGCUCUAGACAUCAGUCAACUAUGAAGUCAACUGCUAUUCUUAGCAGGCAC_CGUAU ...................................................(((((((..(((((.........)))))...)))))))...((((((....)))))).......... (-11.19 = -11.30 + 0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:11:54 2011