
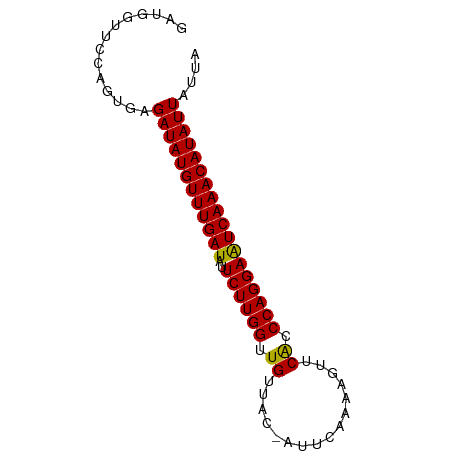
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,564,908 – 8,564,985 |
| Length | 77 |
| Max. P | 0.997168 |

| Location | 8,564,908 – 8,564,985 |
|---|---|
| Length | 77 |
| Sequences | 15 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 90.57 |
| Shannon entropy | 0.22560 |
| G+C content | 0.31812 |
| Mean single sequence MFE | -20.76 |
| Consensus MFE | -19.29 |
| Energy contribution | -19.16 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.997168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8564908 77 + 24543557 GAUGGUUCCAGUGAGAUAUGUUUGAUAUUCUUGGUUGUUUC-AUUCAAAAGUUCACCCAGGAAUCAAACAUAUUAUUA ..............((((((((((((..((((((.((...(-........)..)).)))))))))))))))))).... ( -19.70, z-score = -2.40, R) >droSim1.chr3L 8005922 77 + 22553184 GAUGGUUCCAGUGAGAUAUGUUUGAUAUUCUUGGUUGUAUC-AUUCAAAAGUUCACCCAGGAAUCAAACAUAUUAUUA ..............((((((((((((..((((((.((...(-........)..)).)))))))))))))))))).... ( -19.70, z-score = -2.34, R) >droSec1.super_0 844160 77 + 21120651 GAUGGUUCCAGUGAGAUAUGUUUGAUAUUCUUGGUUGUUUC-AUUCAAAAGUUCACCCAGGAAUCAAACAUAUUAUUA ..............((((((((((((..((((((.((...(-........)..)).)))))))))))))))))).... ( -19.70, z-score = -2.40, R) >droYak2.chr3L 20848865 77 - 24197627 GAUGGUUCCAGUGAGAUAUGUUUGAUAUUCUUGGUUGUUUC-AUUCAAAAGUUCACCCAGGAAUCAAACAUAUUAUUA ..............((((((((((((..((((((.((...(-........)..)).)))))))))))))))))).... ( -19.70, z-score = -2.40, R) >droEre2.scaffold_4784 20279598 77 - 25762168 GAUGGUUCCAGUGAGAUAUGUUUGAUAUUCUUGGUUGUUUC-AUUCAAAAGUUCACCCAGGAAUCAAACAUAUUAUUA ..............((((((((((((..((((((.((...(-........)..)).)))))))))))))))))).... ( -19.70, z-score = -2.40, R) >droAna3.scaffold_13337 15218938 77 - 23293914 GAUGGUUCCAGUGAGAUAUGUUUGAUAUUCUUGGUUGUUAC-AUUCAAAAGUUCACCCAGGAAUCAAACAUAUUAUUA ..............((((((((((((..((((((.((..((-........)).)).)))))))))))))))))).... ( -20.80, z-score = -2.78, R) >dp4.chrXR_group3a 1321117 77 - 1468910 GGGGAUUCCAGUGAGAUAUGUUUGAUAUUCUUGGUUGUUAC-AUUCAAGAGUUCACCCAGGAAUCAAACAUAUUAUUA .((....)).....((((((((((((..((((((.((..((-........)).)).)))))))))))))))))).... ( -21.40, z-score = -2.40, R) >droPer1.super_23 1511572 77 - 1662726 GGGGAUUCCAGUGAGAUAUGUUUGAUAUUCUUGGUUGUUAC-AUUCAAGAGUUCACCCAGGAAUCAAACAUAUUAUUA .((....)).....((((((((((((..((((((.((..((-........)).)).)))))))))))))))))).... ( -21.40, z-score = -2.40, R) >droWil1.scaffold_180949 5214353 77 + 6375548 GAUGGUCCCAGUGAGAUAUGUUUGAUAUUCUUGGUUGUUAC-AUUCAAAAGUUCACCCAGGAAUCAAACAUAUUAUUA ..............((((((((((((..((((((.((..((-........)).)).)))))))))))))))))).... ( -20.80, z-score = -2.65, R) >droVir3.scaffold_13049 6215374 77 + 25233164 AAUGAUUCCAGUAAGAUAUGUUUGAUAUUCUUGGUUGUUAC-AUUCAAAAGUUCACCCAGGAAUCAAACAUAUUAUUA ..............((((((((((((..((((((.((..((-........)).)).)))))))))))))))))).... ( -20.80, z-score = -3.77, R) >droMoj3.scaffold_6680 4536922 77 + 24764193 AACGAUUCCAGUGAGAUAUGUUUGAUAUUCUUGGUUGUUAC-AUUCAAAAGUUCACCCAGGAAUCAAACAUAUUAUUA ..............((((((((((((..((((((.((..((-........)).)).)))))))))))))))))).... ( -20.80, z-score = -3.57, R) >droGri2.scaffold_15072 36315 77 + 214506 AAUGAUUCCAGUGAGAUAUGUUUGAUAUUCUUGGUUGUUAC-AUUCAAAAGUUCACCCAGGAAUCAAACAUAUUAUUA ..............((((((((((((..((((((.((..((-........)).)).)))))))))))))))))).... ( -20.80, z-score = -3.23, R) >anoGam1.chr2L 2608463 78 - 48795086 GACAGUUUCGGUGAGAUAUGUUUGAUAUUCUUGGUUGUUAAGAUUUCAAUUAUCACCCAGGAAUCAAACAUAUUAUUA ..............((((((((((((..((((((.((.(((........))).)).)))))))))))))))))).... ( -21.40, z-score = -3.34, R) >apiMel3.Group13 8331321 75 + 8929068 UCUGGUUUCCGUAAGAUAUGUUUGAUAUUCUUGGUUGUUU---UUUAAAGAAUCGACCAGGAAUCAAACAUAUUAUUA ..............((((((((((((..(((((((((.((---(.....))).))))))))))))))))))))).... ( -24.80, z-score = -5.18, R) >triCas2.ChLG7 16420606 73 - 17478683 GUGAGUUGCUAUAAGAUAUGUUUGAUAUUCUUGGUUGUAUU--UUUAA---UUCACCCAGGAGUCAAACAUAUUAUUA ..............((((((((((((..((((((.((....--.....---..)).)))))))))))))))))).... ( -19.90, z-score = -3.69, R) >consensus GAUGGUUCCAGUGAGAUAUGUUUGAUAUUCUUGGUUGUUAC_AUUCAAAAGUUCACCCAGGAAUCAAACAUAUUAUUA ..............((((((((((((..((((((.((................)).)))))))))))))))))).... (-19.29 = -19.16 + -0.12)


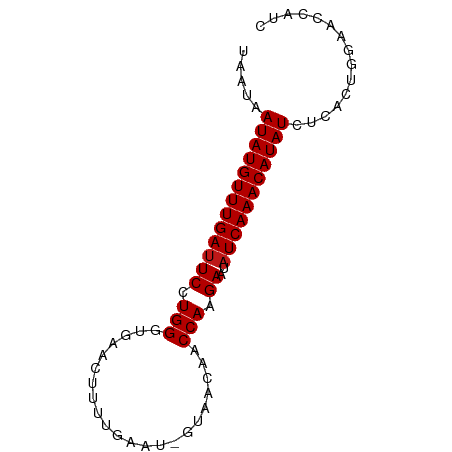
| Location | 8,564,908 – 8,564,985 |
|---|---|
| Length | 77 |
| Sequences | 15 |
| Columns | 78 |
| Reading direction | reverse |
| Mean pairwise identity | 90.57 |
| Shannon entropy | 0.22560 |
| G+C content | 0.31812 |
| Mean single sequence MFE | -14.29 |
| Consensus MFE | -12.88 |
| Energy contribution | -12.94 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.858586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8564908 77 - 24543557 UAAUAAUAUGUUUGAUUCCUGGGUGAACUUUUGAAU-GAAACAACCAAGAAUAUCAAACAUAUCUCACUGGAACCAUC .....(((((((((((..((.(((....((((....-))))..))).))...)))))))))))............... ( -14.20, z-score = -1.31, R) >droSim1.chr3L 8005922 77 - 22553184 UAAUAAUAUGUUUGAUUCCUGGGUGAACUUUUGAAU-GAUACAACCAAGAAUAUCAAACAUAUCUCACUGGAACCAUC .....(((((((((((..((.(((............-......))).))...)))))))))))............... ( -13.57, z-score = -0.91, R) >droSec1.super_0 844160 77 - 21120651 UAAUAAUAUGUUUGAUUCCUGGGUGAACUUUUGAAU-GAAACAACCAAGAAUAUCAAACAUAUCUCACUGGAACCAUC .....(((((((((((..((.(((....((((....-))))..))).))...)))))))))))............... ( -14.20, z-score = -1.31, R) >droYak2.chr3L 20848865 77 + 24197627 UAAUAAUAUGUUUGAUUCCUGGGUGAACUUUUGAAU-GAAACAACCAAGAAUAUCAAACAUAUCUCACUGGAACCAUC .....(((((((((((..((.(((....((((....-))))..))).))...)))))))))))............... ( -14.20, z-score = -1.31, R) >droEre2.scaffold_4784 20279598 77 + 25762168 UAAUAAUAUGUUUGAUUCCUGGGUGAACUUUUGAAU-GAAACAACCAAGAAUAUCAAACAUAUCUCACUGGAACCAUC .....(((((((((((..((.(((....((((....-))))..))).))...)))))))))))............... ( -14.20, z-score = -1.31, R) >droAna3.scaffold_13337 15218938 77 + 23293914 UAAUAAUAUGUUUGAUUCCUGGGUGAACUUUUGAAU-GUAACAACCAAGAAUAUCAAACAUAUCUCACUGGAACCAUC .....(((((((((((..((.(((..((........-))....))).))...)))))))))))............... ( -14.60, z-score = -1.30, R) >dp4.chrXR_group3a 1321117 77 + 1468910 UAAUAAUAUGUUUGAUUCCUGGGUGAACUCUUGAAU-GUAACAACCAAGAAUAUCAAACAUAUCUCACUGGAAUCCCC .....(((((((((((..((.(((..((........-))....))).))...)))))))))))............... ( -14.60, z-score = -1.30, R) >droPer1.super_23 1511572 77 + 1662726 UAAUAAUAUGUUUGAUUCCUGGGUGAACUCUUGAAU-GUAACAACCAAGAAUAUCAAACAUAUCUCACUGGAAUCCCC .....(((((((((((..((.(((..((........-))....))).))...)))))))))))............... ( -14.60, z-score = -1.30, R) >droWil1.scaffold_180949 5214353 77 - 6375548 UAAUAAUAUGUUUGAUUCCUGGGUGAACUUUUGAAU-GUAACAACCAAGAAUAUCAAACAUAUCUCACUGGGACCAUC ......((((((((((..((.(((..((........-))....))).))...))))))))))(((.....)))..... ( -14.90, z-score = -0.89, R) >droVir3.scaffold_13049 6215374 77 - 25233164 UAAUAAUAUGUUUGAUUCCUGGGUGAACUUUUGAAU-GUAACAACCAAGAAUAUCAAACAUAUCUUACUGGAAUCAUU .....(((((((((((..((.(((..((........-))....))).))...)))))))))))............... ( -14.60, z-score = -1.47, R) >droMoj3.scaffold_6680 4536922 77 - 24764193 UAAUAAUAUGUUUGAUUCCUGGGUGAACUUUUGAAU-GUAACAACCAAGAAUAUCAAACAUAUCUCACUGGAAUCGUU .....(((((((((((..((.(((..((........-))....))).))...)))))))))))............... ( -14.60, z-score = -1.25, R) >droGri2.scaffold_15072 36315 77 - 214506 UAAUAAUAUGUUUGAUUCCUGGGUGAACUUUUGAAU-GUAACAACCAAGAAUAUCAAACAUAUCUCACUGGAAUCAUU .....(((((((((((..((.(((..((........-))....))).))...)))))))))))............... ( -14.60, z-score = -1.25, R) >anoGam1.chr2L 2608463 78 + 48795086 UAAUAAUAUGUUUGAUUCCUGGGUGAUAAUUGAAAUCUUAACAACCAAGAAUAUCAAACAUAUCUCACCGAAACUGUC .....(((((((((((((.(((.((.(((........))).)).))).))..)))))))))))............... ( -14.80, z-score = -2.12, R) >apiMel3.Group13 8331321 75 - 8929068 UAAUAAUAUGUUUGAUUCCUGGUCGAUUCUUUAAA---AAACAACCAAGAAUAUCAAACAUAUCUUACGGAAACCAGA .....(((((((((((((.((((.(..........---...).)))).))..))))))))))).....(....).... ( -14.32, z-score = -2.64, R) >triCas2.ChLG7 16420606 73 + 17478683 UAAUAAUAUGUUUGACUCCUGGGUGAA---UUAAA--AAUACAACCAAGAAUAUCAAACAUAUCUUAUAGCAACUCAC .....((((((((((.((.(((.((.(---(....--.)).)).))).))...))))))))))............... ( -12.30, z-score = -2.45, R) >consensus UAAUAAUAUGUUUGAUUCCUGGGUGAACUUUUGAAU_GUAACAACCAAGAAUAUCAAACAUAUCUCACUGGAACCAUC .....(((((((((((((.(((......................))).))..)))))))))))............... (-12.88 = -12.94 + 0.07)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:11:51 2011