| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,563,331 – 8,563,427 |
| Length | 96 |
| Max. P | 0.954106 |

| Location | 8,563,331 – 8,563,427 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 61.25 |
| Shannon entropy | 0.73875 |
| G+C content | 0.34976 |
| Mean single sequence MFE | -12.69 |
| Consensus MFE | -6.02 |
| Energy contribution | -6.04 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.954106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8563331 96 + 24543557 -CAUUCAUCCGCAUAUACAUAUGUAUGUACAAAUCGAA-AAAAU----AAACCAAAAAUCGAAUCGAAAAAUA--CGCCGAGCAGACGCUUCAAGAUGUUUGCC -......((.((((((((....)))))).....((((.-.....----..........))))...........--.)).))((((((((.....).))))))). ( -14.99, z-score = -1.25, R) >droSec1.super_0 842582 102 + 21120651 -CAUUCAUCCGCAUAUACAUAUGUAUGUACAAAUCGAACAAAAUCAAAAAAAAAAAAAUCGAAUCGAAAAAUAA-CGCCGAGCAGACGCUUCAAGAUGUUUUCC -....((((.((((((((....)))))).....((((.....................))))............-.)).((((....))))...))))...... ( -11.70, z-score = -0.43, R) >droYak2.chr3L 20847318 93 - 24197627 -CAUUCAUCCGCAUAUACAUAUGUAUGUACAAAUCGAACAAUAU-----AAC--GAAAUCGAACC--AAAAUAA-CGCCGAGCAGACGCUUCAAGAUGUUUGCC -......((.((((((((....)))))).....((((.......-----...--....))))...--.......-.)).))((((((((.....).))))))). ( -15.14, z-score = -1.11, R) >droEre2.scaffold_4784 20278053 93 - 25762168 -CAUUCAUCCGCAUAUACAUAUGUAUGUACAAAUCGAAAAAUAU-----AAC--GAAAUCGAACA--AAAAUAA-CGCCGAGCAGACGCUUCAAGAUGUUUGCC -......((.((((((((....)))))).....((((.......-----...--....))))...--.......-.)).))((((((((.....).))))))). ( -15.14, z-score = -1.18, R) >droWil1.scaffold_180949 5212444 74 + 6375548 ---------------CAUUCAUCCAU-UAUCAAAAAAUCAAAAU----------GAAA----UGAAAAUAAAACAUUUCGAGCAGACACUGCAAGAUGUUUGCC ---------------((..((((...-................(----------((((----((.........))))))).((((...))))..))))..)).. ( -12.40, z-score = -2.04, R) >droVir3.scaffold_13049 6213440 84 + 25233164 -----------CAUCAAAAUCAAAAUCGAAUUCGAUAUCGAUACC---------AACCAUAUUUCGAAAACAAACAACAAUGCAGACGCUUCAAGAUGUUUGCC -----------.............((((....)))).((((....---------.........))))..............((((((((.....).))))))). ( -11.62, z-score = -1.47, R) >droGri2.scaffold_15110 18045662 94 - 24565398 GAUCUAAAUACCAUCUACUACACCACAUAUGUAUAUAUAAAAAA----------AAAACUGUUUACAACAACAACCACAAAGCAGACGCUUCAAGAUGUUUGCC ...........(((((..((((.......))))...........----------....((((((...............))))))........)))))...... ( -7.86, z-score = -0.50, R) >consensus _CAUUCAUCCGCAUAUACAUAUGUAUGUACAAAUCGAACAAAAU_____AA___AAAAUCGAAUCGAAAAAUAA_CGCCGAGCAGACGCUUCAAGAUGUUUGCC .................................................................................((((((((.....).))))))). ( -6.02 = -6.04 + 0.02)

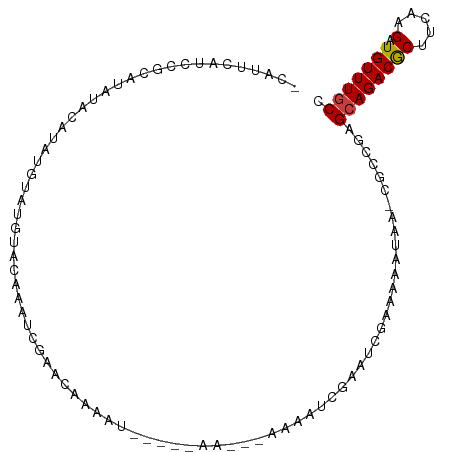
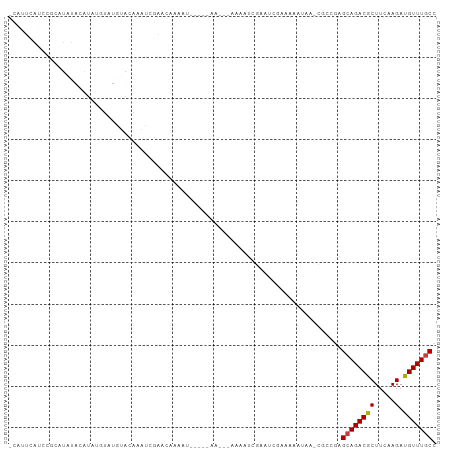
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:11:49 2011