
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,458,342 – 8,458,432 |
| Length | 90 |
| Max. P | 0.536007 |

| Location | 8,458,342 – 8,458,432 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 72.69 |
| Shannon entropy | 0.55121 |
| G+C content | 0.61972 |
| Mean single sequence MFE | -37.22 |
| Consensus MFE | -16.45 |
| Energy contribution | -16.34 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.70 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.536007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
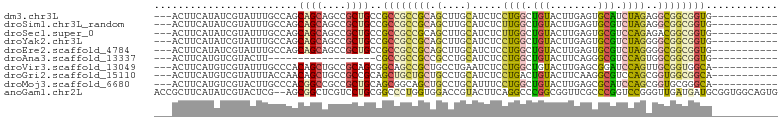
>dm3.chr3L 8458342 90 - 24543557 ---ACUUCAUAUCGUAUUUGCCAGCAGCAGCCGCUGCCGCCGCCGCAGCUUGCAUCUCCUGGCUGUACUUGAGUGCAUCUAGAGGCGGCGGUG----------- ---................(((.(((((....))))).((((((((.....)).....((((.(((((....))))).)))).))))))))).----------- ( -37.60, z-score = -1.61, R) >droSim1.chr3L_random 387288 90 - 1049610 ---ACUUCAUAUCGUAUUUGCCAGCAGCAGCCGCUGCCGCCGCCGCAGCUUGCAUCUCUUGGCUGUACUUGAGUGCGUCUAGAGGCGGCGGUG----------- ---....................(((((....)))))(((((((((.(....)..((((.(((.((((....))))))).)))))))))))))----------- ( -37.40, z-score = -1.37, R) >droSec1.super_0 754267 90 - 21120651 ---ACUUCAUAUCGUAUUUGCCAGCAGCAGCCGCUGCCGCCGCCGCAGCUUGCAUCUCUUGGCUGUACUUGAGUGCGUCCAGAGACGGCGGUG----------- ---....................(((((....)))))(((((((((.....)).(((((.((.(((((....))))).))))))).)))))))----------- ( -36.50, z-score = -1.73, R) >droYak2.chr3L 20758050 90 + 24197627 ---ACUUCAUAUCGUAUUUGCCAGCAGCAGCCGCUGCCGCCGCCGCAGCUUGCAUCUCCUGGCUGUACUUGAGUGCGUCUAGGGGCGGCGGUG----------- ---................(((.(((((....))))).((((((((.....))....(((((.(((((....))))).)))))))))))))).----------- ( -40.00, z-score = -1.64, R) >droEre2.scaffold_4784 20187463 90 + 25762168 ---ACUUCAUAUCGUAUUUGCCAGCAGCAGCCGCUGCCGCCGCCGCAGCUUGCAUCUCCUGGCUGUACUUGAGUGCGUCUAGGGGCGGCGGUG----------- ---................(((.(((((....))))).((((((((.....))....(((((.(((((....))))).)))))))))))))).----------- ( -40.00, z-score = -1.64, R) >droAna3.scaffold_13337 15129604 72 + 23293914 ---ACUUCAUGUCGUACUU------------------CGCCGCCGCCGCCUGCAUCUCCUGGCUGUACUUCAGGGCGUCCAGUGGCGGCGGUG----------- ---................------------------((((((((((((.((.((.((((((.......)))))).)).))))))))))))))----------- ( -33.20, z-score = -3.21, R) >droVir3.scaffold_13049 6094168 90 - 25233164 ---ACUUCAUGUCGUAUUUGCCCACAGCUGCCGCAGCGGCAGCCGCUGCCUGAAUCUCCUGGCUGUACUUGAGCGGAUCCAGUUGCGGUGGCA----------- ---.......................((..(((((((((...((((((((.(......).)))........)))))..)).)))))))..)).----------- ( -33.10, z-score = -0.25, R) >droGri2.scaffold_15110 17944685 90 + 24565398 ---ACUUCAUGUCGUAUUUACCAACAGCUGCCGCCGCAGCUGCUGCUGCCUGCAUCUCCUGACUGUACUUCAAGGCGUCCAGCGGUGGCGGCA----------- ---.......................((((((((((((((....)))(((((((((....)).)))......)))).....))))))))))).----------- ( -34.00, z-score = -1.52, R) >droMoj3.scaffold_6680 4420877 90 - 24764193 ---ACUUCAUGUCGUACUUGCCCACGGCCGCCGCUGCAGCGGCAGCUGCCUGCAUUUCCUGGCUGUACUUGAGCGCAUCCAGCGGUGCGGGCA----------- ---...............(((((.((((.(((((....))))).))))...(((((..((((.((..(....)..)).)))).))))))))))----------- ( -38.10, z-score = -0.47, R) >anoGam1.chr2L 36047059 102 - 48795086 ACCGCUUCAUAUCGUACUCG--AGCGGCUCGUCCUGCGGCCCUGGUGGACCGUACUUCAGGCCCGGCGGUUCGCCCGGUCCGGGUUGAUGAUGCGGUGGCAGUG (((((.((((.....(((((--.((((......))))((((..((((((((((............)))))))))).)))))))))..)))).)))))....... ( -42.30, z-score = 0.12, R) >consensus ___ACUUCAUAUCGUAUUUGCCAGCAGCAGCCGCUGCCGCCGCCGCAGCCUGCAUCUCCUGGCUGUACUUGAGUGCGUCCAGAGGCGGCGGUG___________ ........................((((....))))..((((((((.(....).....((((.(((........))).))))..))))))))............ (-16.45 = -16.34 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:11:42 2011