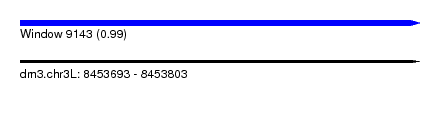
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,453,693 – 8,453,803 |
| Length | 110 |
| Max. P | 0.992876 |

| Location | 8,453,693 – 8,453,803 |
|---|---|
| Length | 110 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 72.07 |
| Shannon entropy | 0.57504 |
| G+C content | 0.45893 |
| Mean single sequence MFE | -29.04 |
| Consensus MFE | -16.23 |
| Energy contribution | -16.79 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.992876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8453693 110 + 24543557 CGUCCCGCUCG-CCGUUGUCGUCAGUCAUUUAGGCGGCGUGUCGUGCCUUAUUGAGUGUAAAUCUGUCGACAAUGCGCAC--AUACACCUCUAUAUAUAGGU-GUGCAUAUAUC ......((..(-(((((((((.(((.(((((((..((((.....))))...))))))).....))).)))))))).))..--..((((((........))))-))))....... ( -36.70, z-score = -2.81, R) >droSim1.chr3L 7911872 110 + 22553184 CAUCCCGCUCG-CCGUUGUCAUCAGUCAUUUACGCGGCGUGUCGUGCCUUAUUGAGUGUAAAUCUGUCGACAAUGCGCAC--AUACACCUCUAUAUAUAGGU-GUGCAUAUAUC ......((..(-((((((((..(((..((((((((((((.....)))).......)))))))))))..))))))).))..--..((((((........))))-))))....... ( -33.51, z-score = -2.48, R) >droYak2.chr3L 20753370 110 - 24197627 CGUCCCGCUCG-CCGUUGUCGUCAGUCAUUUAGGCGGCGUGUCGUGCCUUAUUGAGUGUAAAUCUGUCGACAAUGCGCAC--AUACACCUCUAUAUAUAGGU-GUACAUAUAUC ..........(-(((((((((.(((.(((((((..((((.....))))...))))))).....))).)))))))).))..--.(((((((........))))-)))........ ( -35.90, z-score = -2.89, R) >droEre2.scaffold_4784 20182775 110 - 25762168 CGUCCCGCUCG-CCGUUGUCGUCAGUCAUUUAGGCGGCGUGUCGUGCCUUAUUGAGUGUAAAUCUGUCGACAAUGCGCAC--AUACACCUCUAUAUAUAGGU-GUACAUAUAUC ..........(-(((((((((.(((.(((((((..((((.....))))...))))))).....))).)))))))).))..--.(((((((........))))-)))........ ( -35.90, z-score = -2.89, R) >droAna3.scaffold_13337 15125290 101 - 23293914 -------------CGUCGUCGUCAGUCAUUUAGGCGGCGUGUCGUGCCUUAUUGAGUGUAAAUCUGUCGACAAUGCACACGAAUGUAUAUACACUCCUACCCAGUGUGUAUAUC -------------(((.((((.(((.(((((((..((((.....))))...))))))).....))).)))).))).........(((((((((((.......))))))))))). ( -29.50, z-score = -1.80, R) >dp4.chrXR_group6 9431981 110 + 13314419 UCUCCCUAGCC-CCGCUGUCGUCAGUCAUUUAGGCGGCGUGUCGUGCCUUAUUGAGUGUAAAUCUGUCGACAAUACACAC--AUACAUUCUCAUACAUACGU-AUAUGUAUGUA ...........-....(((((.(((.(((((((..((((.....))))...))))))).....))).)))))......((--((((((....((((....))-)))))))))). ( -28.30, z-score = -1.79, R) >droMoj3.scaffold_6680 4415914 97 + 24764193 CACCCUACGCU-GCCGCGUCGUCAGUCAUUUAGGCGGCGUGUCGUGGCUUAUUGAGUGUAAAUCUGUCGACAAUAUUUUCU-GUUUUAUUUAAUUUUUU--------------- .....((((((-((((((.((((.(((.....)))))))...))))))......))))))........((((........)-)))..............--------------- ( -22.00, z-score = -0.98, R) >droVir3.scaffold_13049 6089526 105 + 25233164 CACCCUACGCC-GCCUCAUCGUCAGUCAUUUAGGCGGCGUGUCGUGCCUUAUUGAGUGUAAAUCUGUCGACAAUAUUUUCU-GUUUUAUUUAAUUUUUCCAUGUUUC------- .....((((((-((((...............)))))))))).((((....((((((((..........((((........)-))).)))))))).....))))....------- ( -20.96, z-score = -1.81, R) >droGri2.scaffold_15110 17940022 91 - 24565398 -------CGCCUGCCUCAUCGUCAGUCAUUUAGGCGGCGUGUCGUGCCUUAUUGAGUGUAAAUCUGUCGACAAUAAUUUUU-UUUUGUUUUCGCUUGCC--------------- -------.((..((....(((.(((.(((((((..((((.....))))...))))))).....))).))).(((((.....-..)))))...))..)).--------------- ( -18.60, z-score = -0.42, R) >consensus CAUCCCGCGCG_CCGUUGUCGUCAGUCAUUUAGGCGGCGUGUCGUGCCUUAUUGAGUGUAAAUCUGUCGACAAUGCGCAC__AUACACCUCUAUAUAUAGGU_GUACAUAUAUC .................((((.(((.(((((((..((((.....))))...))))))).....))).))))........................................... (-16.23 = -16.79 + 0.56)

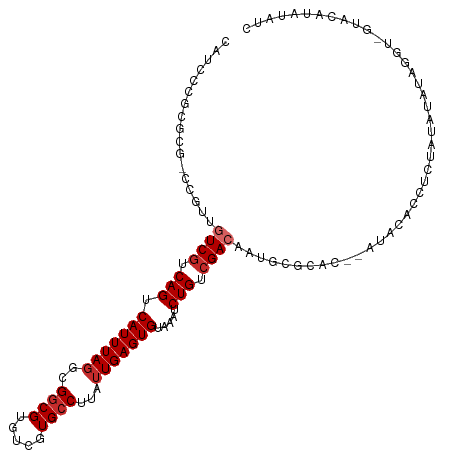
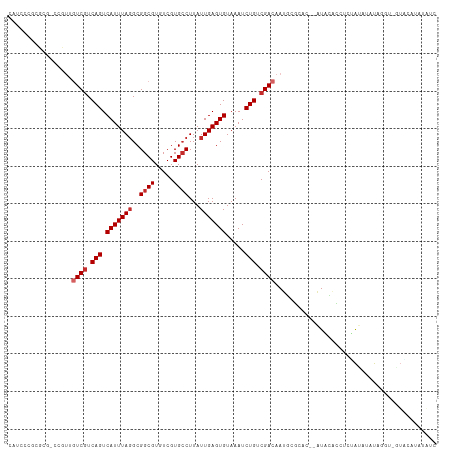
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:11:41 2011