| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,448,011 – 8,448,139 |
| Length | 128 |
| Max. P | 0.992160 |

| Location | 8,448,011 – 8,448,139 |
|---|---|
| Length | 128 |
| Sequences | 4 |
| Columns | 148 |
| Reading direction | forward |
| Mean pairwise identity | 67.77 |
| Shannon entropy | 0.48188 |
| G+C content | 0.37835 |
| Mean single sequence MFE | -32.52 |
| Consensus MFE | -16.92 |
| Energy contribution | -18.29 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.894603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
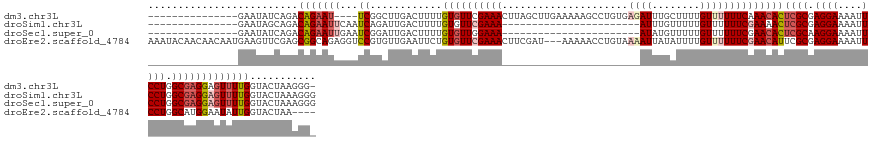
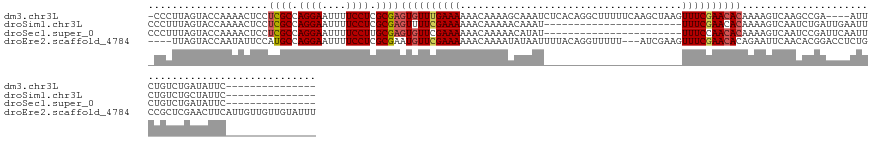
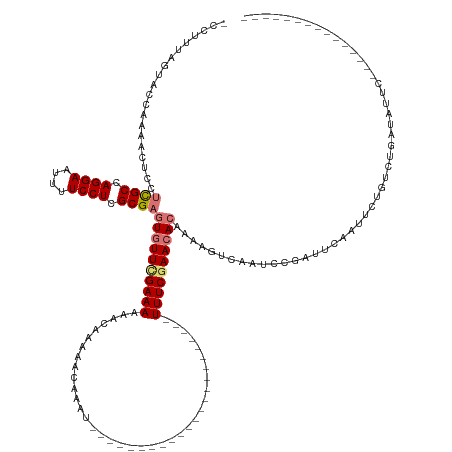
>dm3.chr3L 8448011 128 + 24543557 ---------------GAAUAUCAGACAGAAU----UCGGCUUGACUUUUGUGUUCGAAACUUAGCUUGAAAAAGCCUGUGAGAUUUGCUUUUGUUUUUUCAAACACUCGCGAGGAAAAUUCCUGGCGAGGAGUUUUGGUACUAAGGG- ---------------((((.((.....))))----)).......((((((.(((((((((((.(.((((((((((....(((.....)))..)))))))))).).(((((.((((....)))).)))))))))))))).))))))))- ( -37.30, z-score = -1.86, R) >droSim1.chr3L 7906327 110 + 22553184 ---------------GAAUAGCAGACAGAAUUCAAUCAGAUUGACUUUUGUGUUCGAAA-----------------------AUUUGUUUUUGUUUUUUCGAAAACUCGCGAGGAAAAUUCCUGGCGAGGAGUUUUGGUACUAAAGGG ---------------...(((....((((((((...((((......))))..(((((((-----------------------(..(......)..))))))))..(((((.((((....)))).)))))))))))))...)))..... ( -30.40, z-score = -2.08, R) >droSec1.super_0 743884 110 + 21120651 ---------------GAAUAUCAGACAGAAUUGAAUCGGAUUGACUUUUGUGUUGGAAA-----------------------AUAUGUUUUUGUUUUUUCGAACACUCGCAAGGAAAAUUCCUGGCGAGGAGUUUUGGUACUAAAGGG ---------------..........(((((((...(((((..(((...((((((....)-----------------------))))).....)))..)))))...(((((.((((....)))).))))).)))))))........... ( -28.20, z-score = -1.86, R) >droEre2.scaffold_4784 2172935 141 - 25762168 AAAUACAACAACAAUGAAGUUCGAGCGGCAGAGGUCCGUGUUGAAUUCUGUGUUCGAAACUUCGAU---AAAAACCUGUAAAAUUAUAUUUUGUUUUUUCGAACAUUCGCGAGGAAAAUUCCUGGCAUGGAAUAUUGGUACUAA---- ..............(((((((((((((.(((((.((......)).))))))))))).)))))))..---....(((.(((...........(((((....)))))...((.((((....)))).))......))).))).....---- ( -34.20, z-score = -1.47, R) >consensus _______________GAAUAUCAGACAGAAUUGAAUCGGAUUGACUUUUGUGUUCGAAA_______________________AUUUGUUUUUGUUUUUUCGAACACUCGCGAGGAAAAUUCCUGGCGAGGAGUUUUGGUACUAAAGG_ .........................((((((...................(((((((((.....................................)))))))))(((((.((((....)))).)))))..))))))........... (-16.92 = -18.29 + 1.37)
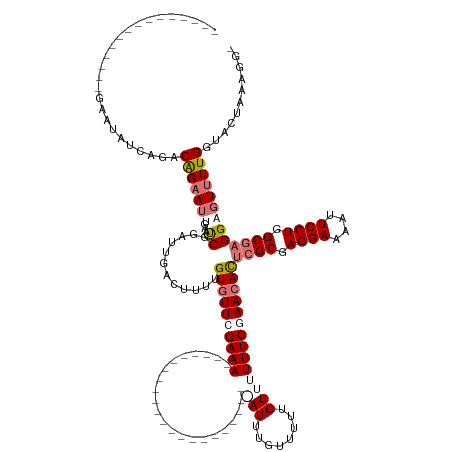
| Location | 8,448,011 – 8,448,139 |
|---|---|
| Length | 128 |
| Sequences | 4 |
| Columns | 148 |
| Reading direction | reverse |
| Mean pairwise identity | 67.77 |
| Shannon entropy | 0.48188 |
| G+C content | 0.37835 |
| Mean single sequence MFE | -24.75 |
| Consensus MFE | -14.83 |
| Energy contribution | -15.52 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.992160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8448011 128 - 24543557 -CCCUUAGUACCAAAACUCCUCGCCAGGAAUUUUCCUCGCGAGUGUUUGAAAAAACAAAAGCAAAUCUCACAGGCUUUUUCAAGCUAAGUUUCGAACACAAAAGUCAAGCCGA----AUUCUGUCUGAUAUUC--------------- -..((((((..........(((((.((((....)))).)))))(((((....)))))(((((...........))))).....))))))..............((((..(.(.----...).)..))))....--------------- ( -27.50, z-score = -2.00, R) >droSim1.chr3L 7906327 110 - 22553184 CCCUUUAGUACCAAAACUCCUCGCCAGGAAUUUUCCUCGCGAGUUUUCGAAAAAACAAAAACAAAU-----------------------UUUCGAACACAAAAGUCAAUCUGAUUGAAUUCUGUCUGCUAUUC--------------- .....(((((.........(((((.((((....)))).)))))..((((((((............)-----------------------))))))).(((.((.(((((...))))).)).))).)))))...--------------- ( -21.50, z-score = -2.57, R) >droSec1.super_0 743884 110 - 21120651 CCCUUUAGUACCAAAACUCCUCGCCAGGAAUUUUCCUUGCGAGUGUUCGAAAAAACAAAAACAUAU-----------------------UUUCCAACACAAAAGUCAAUCCGAUUCAAUUCUGUCUGAUAUUC--------------- ..((((..............((((.((((....)))).))))(((((.(((((............)-----------------------)))).))))).))))...(((.(((........))).)))....--------------- ( -20.20, z-score = -2.90, R) >droEre2.scaffold_4784 2172935 141 + 25762168 ----UUAGUACCAAUAUUCCAUGCCAGGAAUUUUCCUCGCGAAUGUUCGAAAAAACAAAAUAUAAUUUUACAGGUUUUU---AUCGAAGUUUCGAACACAGAAUUCAACACGGACCUCUGCCGCUCGAACUUCAUUGUUGUUGUAUUU ----......(.(((((((...((.((((....)))).))))))))).)........(((((((((...((((((....---)))((((((.(((.(.((((..((......))..))))..).)))))))))..))).))))))))) ( -29.80, z-score = -1.76, R) >consensus _CCUUUAGUACCAAAACUCCUCGCCAGGAAUUUUCCUCGCGAGUGUUCGAAAAAACAAAAACAAAU_______________________UUUCGAACACAAAAGUCAAUCCGAUUCAAUUCUGUCUGAUAUUC_______________ ....................((((.((((....)))).))))((((((((((.....................................))))))))))................................................. (-14.83 = -15.52 + 0.69)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:11:40 2011