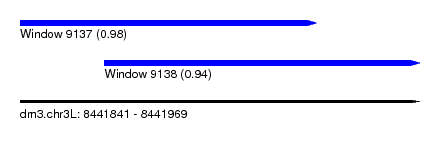
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,441,841 – 8,441,969 |
| Length | 128 |
| Max. P | 0.981847 |

| Location | 8,441,841 – 8,441,936 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 60.44 |
| Shannon entropy | 0.75138 |
| G+C content | 0.42753 |
| Mean single sequence MFE | -20.88 |
| Consensus MFE | -11.29 |
| Energy contribution | -11.93 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.981847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
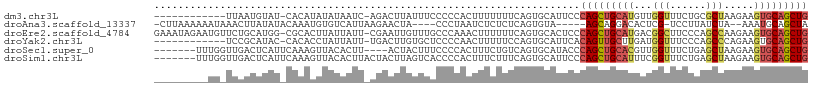
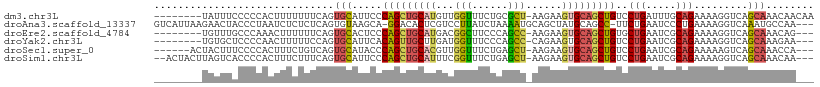
>dm3.chr3L 8441841 95 + 24543557 ------------UUAAUGUAU-CACAUAUAUAAUC-AGACUUAUUUCCCCCACUUUUUUUCAGUGCAUUCCCAGCUGCAUGUUGGUUUCUGCGCUAAGAAGUGCAGCUG ------------...((((((-...))))))....-..............((((((((...((((((...(((((.....)))))....))))))))))))))...... ( -19.80, z-score = -1.56, R) >droAna3.scaffold_13337 15114300 96 - 23293914 -CUUAAAAAAUAAACUUAUAUACAAAUGUGUCAUUAAGAACUA----CCCUAAUCUCUCUCAGUGUA-----AGCAGGACACUCG-UCCUUAUCUA--AAAUGCAGCUA -.............((((.((((....))))...)))).....----..............((((((-----((.(((((....)-))))...)).--...)))).)). ( -9.90, z-score = 0.11, R) >droEre2.scaffold_4784 20171249 107 - 25762168 GAAAUAGAAUGUUCUGCAUGG-CGCACUUAUUAUU-CGAAUUGUUUGCCCAAACUUUUUUCAGUGCACUCCCAGCUGCAUGACGGCUUCCCAGCCAAGAAGUGCAGCUG ....((((....))))...((-.(((((.......-.(((..((((....))))....)))))))).))..(((((((((...((((....)))).....))))))))) ( -27.40, z-score = -0.77, R) >droYak2.chr3L 20741526 95 - 24197627 ------------UCCGCAUAC-CACACCUAUUAUU-UGACUUGUGCUCCCCAACUUUUUCCAGUGCAUUCACAGUUGCUUGAUGGUUUCCCAGCCCAGAAGUGCAGCUG ------------.........-.............-(((..((..((..............))..)).)))(((((((((..(((.........)))..)).))))))) ( -15.94, z-score = 0.04, R) >droSec1.super_0 737770 98 + 21120651 -------UUUGGUUGACUCAUUCAAAGUUACACUU----ACUACUUUCCCCACUUUCUGUCAGUGCAUACCCAGCUGCACGUUGGUUUCUGAGCUAAGAAGUGCAGCUG -------...((((((((.......)))))((((.----((.................)).))))...)))(((((((((.((((((....))))))...))))))))) ( -24.83, z-score = -1.26, R) >droSim1.chr3L 7900138 102 + 22553184 -------UUUGGUUGACUCAUUCAAAGUUACACUUACUACUUAGUCACCCCACUUUCUUUCAGUGCAUUCCCAGCUGCAUUUCGGUUUCUGAGCUAAGAAGUGCAGCUG -------...((.(((((......(((.....))).......))))).))((((.......))))......((((((((((((((((....))))..)))))))))))) ( -27.42, z-score = -2.82, R) >consensus _________UG_UUCACAUAU_CAAACUUAUAAUU_AGAAUUAUUUUCCCCAAUUUCUUUCAGUGCAUUCCCAGCUGCAUGUUGGUUUCUGAGCUAAGAAGUGCAGCUG .......................................................................(((((((((...(((......))).....))))))))) (-11.29 = -11.93 + 0.64)
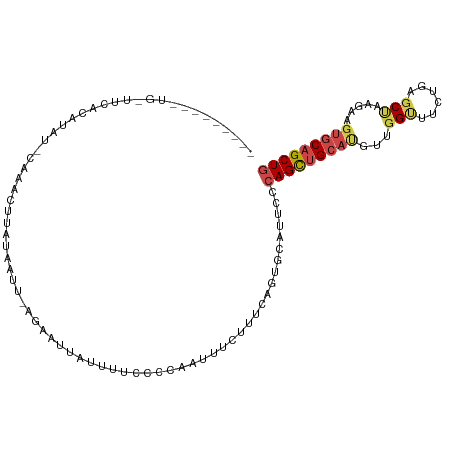
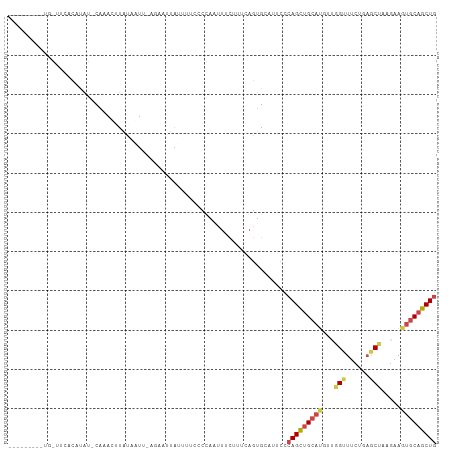
| Location | 8,441,868 – 8,441,969 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 70.26 |
| Shannon entropy | 0.54582 |
| G+C content | 0.46928 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -14.22 |
| Energy contribution | -14.43 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.939286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
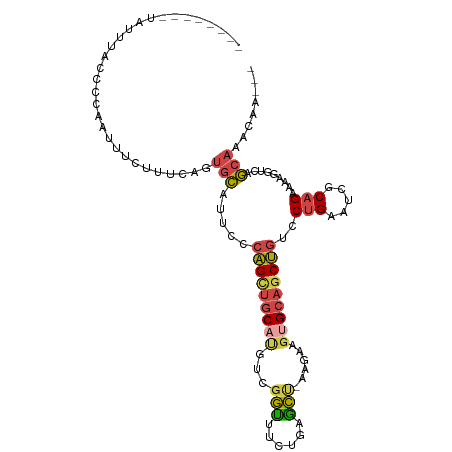
>dm3.chr3L 8441868 101 + 24543557 --------UAUUUCCCCCACUUUUUUUCAGUGCAUUCCCAGCUGCAUGUUGGUUUCUGCGCU-AAGAAGUGCAGCUGUCCUGAUUUGCAGAAAAGGUCAGCAAACAACAA --------.........((((((((...((((((...(((((.....)))))....))))))-))))))))..((((.(((............))).))))......... ( -29.90, z-score = -2.29, R) >droAna3.scaffold_13337 15114328 105 - 23293914 GUCAUUAAGAACUACCCUAAUCUCUCUCAGUGUAAGCA-GGACACUCGUCCUUAUCUAAAAUGCAGCUAUGCAGCC-UUCUGAAUCCCUGAAAAGGUCAAAUGCCAA--- (.((((....................((((.....(((-((((....))))).........((((....)))))).-..))))...(((....)))...)))).)..--- ( -19.50, z-score = -0.97, R) >droEre2.scaffold_4784 20171288 98 - 25762168 --------UGUUUGCCCAAACUUUUUUCAGUGCACUCCCAGCUGCAUGACGGCUUCCCAGCC-AAGAAGUGCAGCUGUGCUGAAUCGCAGAAAAGGUCAGCAAACAG--- --------(((((((....((((((((...........(((((((((...((((....))))-.....)))))))))(((......)))))))))))..))))))).--- ( -34.40, z-score = -2.49, R) >droYak2.chr3L 20741553 98 - 24197627 --------UGUGCUCCCCAACUUUUUCCAGUGCAUUCACAGUUGCUUGAUGGUUUCCCAGCC-CAGAAGUGCAGCUGUCCUGAAUCGCAGAAAAGGUCAGCAAAGAA--- --------..((((.....((((((((..(((.((((((((((((((..(((.........)-))..)).))))))))...))))))).)))))))).)))).....--- ( -27.20, z-score = -1.33, R) >droSec1.super_0 737798 100 + 21120651 ------ACUACUUUCCCCACUUUCUGUCAGUGCAUACCCAGCUGCACGUUGGUUUCUGAGCU-AAGAAGUGCAGCUGUCCUGAAUCGCAGAAAAAGUCAGCAAACCA--- ------..............(((((((.(...((....(((((((((.((((((....))))-))...)))))))))...))..).)))))))..............--- ( -26.70, z-score = -1.42, R) >droSim1.chr3L 7900166 104 + 22553184 --ACUACUUAGUCACCCCACUUUCUUUCAGUGCAUUCCCAGCUGCAUUUCGGUUUCUGAGCU-AAGAAGUGCAGCUGUCCUGAAUCGCAGAAAAGGUCAGCAAACAA--- --........((.(((....(((((....(((.((((.((((((((((((((((....))))-..))))))))))))....)))))))))))).)))..))......--- ( -27.40, z-score = -1.52, R) >consensus ________UAUUUACCCCAAUUUCUUUCAGUGCAUUCCCAGCUGCAUGUCGGUUUCUGAGCU_AAGAAGUGCAGCUGUCCUGAAUCGCAGAAAAGGUCAGCAAACAA___ ..............................(((.....(((((((((...(((......)))......)))))))))..(((.....))).........)))........ (-14.22 = -14.43 + 0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:11:37 2011