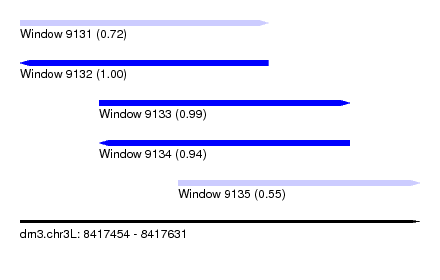
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,417,454 – 8,417,631 |
| Length | 177 |
| Max. P | 0.997089 |

| Location | 8,417,454 – 8,417,564 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 94.55 |
| Shannon entropy | 0.07513 |
| G+C content | 0.43030 |
| Mean single sequence MFE | -28.44 |
| Consensus MFE | -22.15 |
| Energy contribution | -21.93 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.715919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8417454 110 + 24543557 GGGGAGAGUUCGCGCCUCUGCAGGAAAAAUACGUACAUACAAAGUAUAUCCUCAAAAGGAAAGAAAUACUUUGUAUGGCGCAUCAAUUUCCAACACAAGUCGCAUUUAAG (.((((.((....)))))).).(((((.((.(((.((((((((((((.((((....)))).....))))))))))))))).))...)))))................... ( -28.00, z-score = -2.95, R) >droSec1.super_0 713512 110 + 21120651 GGGGAGAGUUCGCGCCUCUGCAGGAAAAAUACGUACAUACAGAGUACAUCCCCGAAAGGAAACAAAUACUUUGUAUGGCGCAUCAAUUUGCAACACAAGUCCCAUUUAAG .(((((((.......)))((((((.......(((.(((((((((((.....((....)).......))))))))))))))......)))))).......))))....... ( -30.12, z-score = -2.76, R) >droSim1.chr3L 7876533 110 + 22553184 GGGGAGAGUUCGCGCCUCUGCAGGAAAAAUACGUACAUACAGAGUACUUCCCUGAAAGGAAAGAAAUACUUUGUAUGGCGCAUCAAUUUGCAACACAAGUCGCAUUUAAG ((((...(....).))))(((...........((.(((((((((((.(((.((....))...))).)))))))))))))(((......)))..........)))...... ( -27.20, z-score = -1.31, R) >consensus GGGGAGAGUUCGCGCCUCUGCAGGAAAAAUACGUACAUACAGAGUACAUCCCCGAAAGGAAAGAAAUACUUUGUAUGGCGCAUCAAUUUGCAACACAAGUCGCAUUUAAG (..(..((((.(((((((.....))...........((((((((((..(((......)))......)))))))))))))))...))))..)..)................ (-22.15 = -21.93 + -0.22)

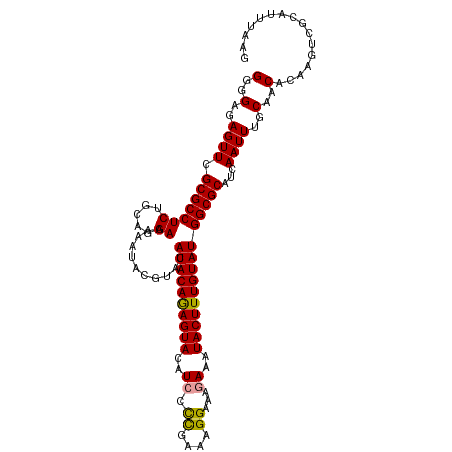
| Location | 8,417,454 – 8,417,564 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 94.55 |
| Shannon entropy | 0.07513 |
| G+C content | 0.43030 |
| Mean single sequence MFE | -35.93 |
| Consensus MFE | -27.51 |
| Energy contribution | -27.41 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.36 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.997089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8417454 110 - 24543557 CUUAAAUGCGACUUGUGUUGGAAAUUGAUGCGCCAUACAAAGUAUUUCUUUCCUUUUGAGGAUAUACUUUGUAUGUACGUAUUUUUCCUGCAGAGGCGCGAACUCUCCCC ......((((.(((.(((.(((((..((((((.((((((((((((.(((((......))))).))))))))))))..))))))))))).))).))))))).......... ( -40.30, z-score = -6.59, R) >droSec1.super_0 713512 110 - 21120651 CUUAAAUGGGACUUGUGUUGCAAAUUGAUGCGCCAUACAAAGUAUUUGUUUCCUUUCGGGGAUGUACUCUGUAUGUACGUAUUUUUCCUGCAGAGGCGCGAACUCUCCCC .......((((.((((((((((....((((((.((((((.(((((..((..((....))..))))))).))))))..)))))).....))))...))))))....)))). ( -34.10, z-score = -2.91, R) >droSim1.chr3L 7876533 110 - 22553184 CUUAAAUGCGACUUGUGUUGCAAAUUGAUGCGCCAUACAAAGUAUUUCUUUCCUUUCAGGGAAGUACUCUGUAUGUACGUAUUUUUCCUGCAGAGGCGCGAACUCUCCCC ......((((((....))))))......(((((((((((.(((((((((((......))))))))))).)))))....(((.......)))...)))))).......... ( -33.40, z-score = -3.58, R) >consensus CUUAAAUGCGACUUGUGUUGCAAAUUGAUGCGCCAUACAAAGUAUUUCUUUCCUUUCGGGGAUGUACUCUGUAUGUACGUAUUUUUCCUGCAGAGGCGCGAACUCUCCCC ......((((.(((.(((........((((((.((((((.(((((.....((((....)))).))))).))))))..))))))......))).))))))).......... (-27.51 = -27.41 + -0.11)


| Location | 8,417,489 – 8,417,600 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 90.27 |
| Shannon entropy | 0.13237 |
| G+C content | 0.35410 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -19.87 |
| Energy contribution | -19.77 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.989037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8417489 111 + 24543557 CAUACAAAGUAUAUCCUCAAAAGGAAAGAAAUACUUUGUAUGGCGCAUCAAUUUCCAACACAAGUCGCAUUUAAGGCGAUAGUUGAUCGAUCAUCUAAGACUUAAAAUUAA ((((((((((((.((((....)))).....)))))))))))).((.(((((((..........(((((.......))))))))))))))...................... ( -25.60, z-score = -3.72, R) >droSec1.super_0 713547 107 + 21120651 CAUACAGAGUACAUCCCCGAAAGGAAACAAAUACUUUGUAUGGCGCAUCAAUUUGCAACACAAGUCCCAUUUAAGGCGAUAGUUGAUC----AUCAAAGACUUUAAAUUAA (((((((((((.....((....)).......)))))))))))..(((......))).....(((((((......)).(((.....)))----......)))))........ ( -23.80, z-score = -3.06, R) >droSim1.chr3L 7876568 107 + 22553184 CAUACAGAGUACUUCCCUGAAAGGAAAGAAAUACUUUGUAUGGCGCAUCAAUUUGCAACACAAGUCGCAUUUAAGGCGAUAGUCGAUC----AUCUAAGACUUUAAAUUAA (((((((((((.(((.((....))...))).)))))))))))..(((......))).......(((((.......)))))((((....----......))))......... ( -26.80, z-score = -3.16, R) >consensus CAUACAGAGUACAUCCCCGAAAGGAAAGAAAUACUUUGUAUGGCGCAUCAAUUUGCAACACAAGUCGCAUUUAAGGCGAUAGUUGAUC____AUCUAAGACUUUAAAUUAA (((((((((((..(((......)))......)))))))))))..(((......))).....(((((........(((....)))(((.....)))...)))))........ (-19.87 = -19.77 + -0.11)

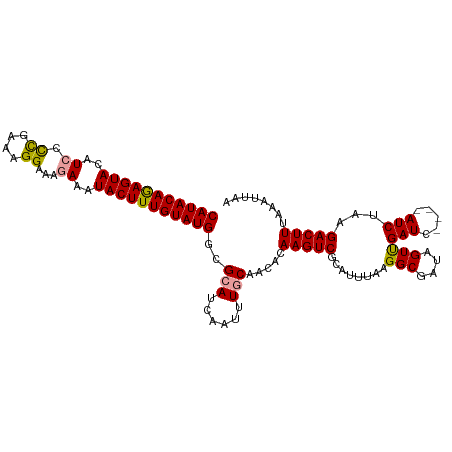
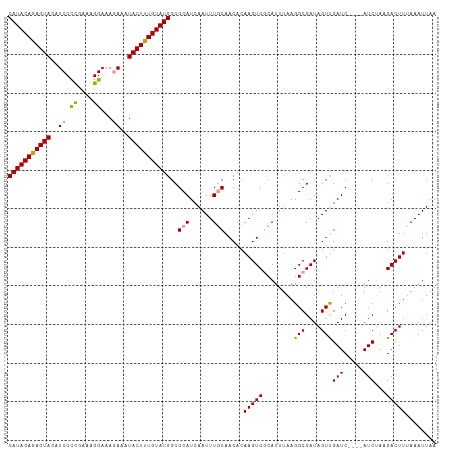
| Location | 8,417,489 – 8,417,600 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 90.27 |
| Shannon entropy | 0.13237 |
| G+C content | 0.35410 |
| Mean single sequence MFE | -26.97 |
| Consensus MFE | -20.24 |
| Energy contribution | -20.47 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.935066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8417489 111 - 24543557 UUAAUUUUAAGUCUUAGAUGAUCGAUCAACUAUCGCCUUAAAUGCGACUUGUGUUGGAAAUUGAUGCGCCAUACAAAGUAUUUCUUUCCUUUUGAGGAUAUACUUUGUAUG ((((((((..(((......)))....((((..((((.......)))).....)))))))))))).....((((((((((((.(((((......))))).)))))))))))) ( -26.90, z-score = -3.05, R) >droSec1.super_0 713547 107 - 21120651 UUAAUUUAAAGUCUUUGAU----GAUCAACUAUCGCCUUAAAUGGGACUUGUGUUGCAAAUUGAUGCGCCAUACAAAGUAUUUGUUUCCUUUCGGGGAUGUACUCUGUAUG ..(((...(((((...(((----(......)))).((......)))))))..)))(((......)))..((((((.(((((..((..((....))..))))))).)))))) ( -24.40, z-score = -1.61, R) >droSim1.chr3L 7876568 107 - 22553184 UUAAUUUAAAGUCUUAGAU----GAUCGACUAUCGCCUUAAAUGCGACUUGUGUUGCAAAUUGAUGCGCCAUACAAAGUAUUUCUUUCCUUUCAGGGAAGUACUCUGUAUG .........((((......----....))))..(((.((((.((((((....))))))..)))).))).((((((.(((((((((((......))))))))))).)))))) ( -29.60, z-score = -3.69, R) >consensus UUAAUUUAAAGUCUUAGAU____GAUCAACUAUCGCCUUAAAUGCGACUUGUGUUGCAAAUUGAUGCGCCAUACAAAGUAUUUCUUUCCUUUCGGGGAUGUACUCUGUAUG .................................(((.((((.((((((....))))))..)))).))).((((((.(((((.....((((....)))).))))).)))))) (-20.24 = -20.47 + 0.23)

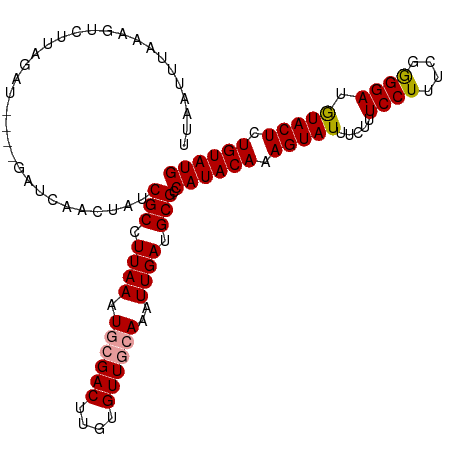
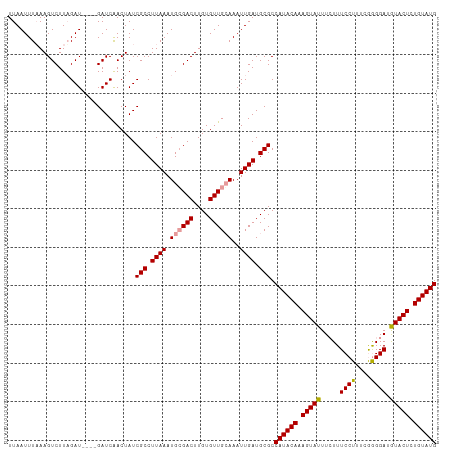
| Location | 8,417,524 – 8,417,631 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 90.85 |
| Shannon entropy | 0.12638 |
| G+C content | 0.33209 |
| Mean single sequence MFE | -21.70 |
| Consensus MFE | -18.83 |
| Energy contribution | -18.50 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.554438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8417524 107 + 24543557 UUGUAUGGCGCAUCAAUUUCCAACACAAGUCGCAUUUAAGGCGAUAGUUGAUCGAUCAUCUAAGACUUAAAAUUAAGUUGAUGUAUAUGAUACAAGAAAUAGUGCCU ......(((((....(((((((((....(((((.......))))).))))((((((((((...((((((....)))))))))).)).))))....))))).))))). ( -24.30, z-score = -1.79, R) >droSec1.super_0 713582 103 + 21120651 UUGUAUGGCGCAUCAAUUUGCAACACAAGUCCCAUUUAAGGCGAUAGUUGAUC----AUCAAAGACUUUAAAUUAAAUUGAUGUAUAAGAUACGAAAAAUAAUGCCU ((((((.(..((((((((((......(((((((......)).(((.....)))----......))))).....))))))))))..)...))))))............ ( -18.50, z-score = -0.91, R) >droSim1.chr3L 7876603 103 + 22553184 UUGUAUGGCGCAUCAAUUUGCAACACAAGUCGCAUUUAAGGCGAUAGUCGAUC----AUCUAAGACUUUAAAUUAAAUUGAUGUAUAAGAUACGAAAAAUAUUGCCU ......((((((((((((((........(((((.......)))))((((....----......))))......)))))))))))....((((.......))))))). ( -22.30, z-score = -1.69, R) >consensus UUGUAUGGCGCAUCAAUUUGCAACACAAGUCGCAUUUAAGGCGAUAGUUGAUC____AUCUAAGACUUUAAAUUAAAUUGAUGUAUAAGAUACGAAAAAUAAUGCCU ((((((.(..((((((((((......(((((........(((....)))(((.....)))...))))).....))))))))))..)...))))))............ (-18.83 = -18.50 + -0.33)

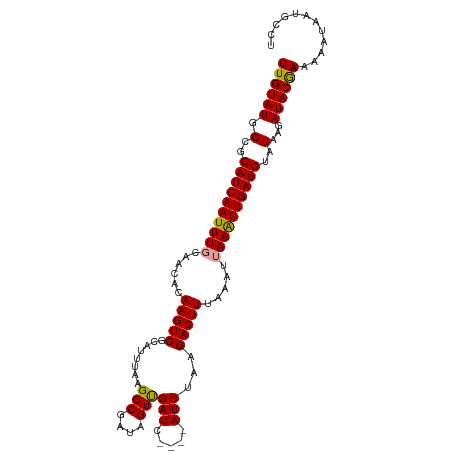
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:11:35 2011