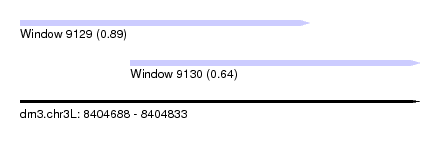
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,404,688 – 8,404,833 |
| Length | 145 |
| Max. P | 0.893005 |

| Location | 8,404,688 – 8,404,793 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 83.89 |
| Shannon entropy | 0.28930 |
| G+C content | 0.41502 |
| Mean single sequence MFE | -29.46 |
| Consensus MFE | -25.04 |
| Energy contribution | -25.28 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.893005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
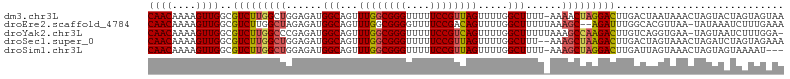
>dm3.chr3L 8404688 105 + 24543557 CAACAAAAGUUGGCGUCUUGGCUGGAGAUGGCAGUUUGGCGGGUUUUUCCGUUAGUUUUGGCUUUU-AAAACUAGGACUUGACUAAUAAACUAGUACUAGUAGUAA .......(((..(.(((((((...((((.(.(((.((((((((....))))))))..))).)))))-....))))))))..))).....((((....))))..... ( -26.70, z-score = -0.73, R) >droEre2.scaffold_4784 20134208 103 - 25762168 CAACAAAAGUUGGCGUCUUGGCUAGAGAUGGCAGUUUGGCGGGGUUUUCCGACAGUUUUGGCUUUUUAAAGC--AGAUUUGGCACGUUAA-UAUAAAUCUUUGAAA ...((((((((((...(((.((((((........)))))).)))....)))))..)))))((((....))))--(((((((.........-..)))))))...... ( -21.50, z-score = 0.62, R) >droYak2.chr3L 20703827 104 - 24197627 CAACAAAAGUUGGCGUCUUGGCCCGAGAUGGCAGUUUGGCGGGUUUUUCCGUCAGUUUUGGCUUUUUAAAGCCAAGACUUGUCAGGUGAA-UAGUAAUCUUUGGA- ((((....))))((......))(((((.((((((..(((((((....)))))))((((((((((....))))))))))))))))(((...-.....)))))))).- ( -33.90, z-score = -2.18, R) >droSec1.super_0 700744 104 + 21120651 CAACAAAAGUUGGCGUCUUGGCUGGAGAUGGCAGUUUGGCGGGUUUUUCCGUUAGUUUUGGCUUU--AAAGCUAAGACUUGACUAGUAAACUAGAUCUAGUAGAAA ((((....))))(((((((((((..(((.(.(((.((((((((....))))))))..))).))))--..)))))))))..((((((....)))).))..))..... ( -33.20, z-score = -2.56, R) >droSim1.chr3L 7865131 102 + 22553184 CAACAAAAGUUGGCGUCUUGGCUGGAGAUGGCAGUUUGGCGGGUUUUUCCGUUAGUUUUGGCUUUU-AAAGCUAGGACUUGAUUAGUAAACUAGUAGUAAAAU--- ((((....))))(((((((((((..(((.(.(((.((((((((....))))))))..))).).)))-..)))))))))...(((((....))))).)).....--- ( -32.00, z-score = -3.03, R) >consensus CAACAAAAGUUGGCGUCUUGGCUGGAGAUGGCAGUUUGGCGGGUUUUUCCGUUAGUUUUGGCUUUU_AAAGCUAAGACUUGACUAGUAAACUAGUAAUAGUAGAAA ((((....))))..(((((((((......(((...((((((((....)))))))).....)))......)))))))))............................ (-25.04 = -25.28 + 0.24)
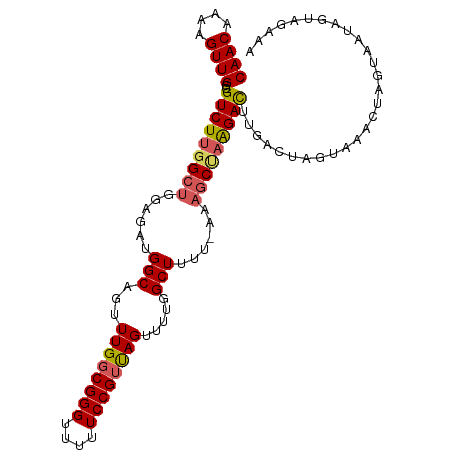
| Location | 8,404,728 – 8,404,833 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 74.30 |
| Shannon entropy | 0.46138 |
| G+C content | 0.29615 |
| Mean single sequence MFE | -18.74 |
| Consensus MFE | -11.90 |
| Energy contribution | -11.98 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.637744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8404728 105 + 24543557 GGGUUUUUCCGUUAGUUUUGGCUUUU-AAAACUAGGACUUGACUAAUAAACUAGUACUAGUAGUAAAAACAAUUUAUUUAAAACAUAGCCUCAUUAUCGUUUACCU ((((.....((.((((...(((((((-((((((((......((((......)))).))))).((....))......))))))....))))..)))).))...)))) ( -16.00, z-score = 0.13, R) >droEre2.scaffold_4784 20134248 101 - 25762168 GGGGUUUUCCGACAGUUUUGGCUUUUUAAAGC--AGAUUUGGCACGUUAA-UAUAAAUCUUUG--AAAAACAGUAAAUAAAAACAUAGCCUAAUUAUCAUUUAACU ..((....))((.((((..((((((((((...--(((((((.........-..))))))))))--))))...((........))...))).)))).))........ ( -11.80, z-score = 1.01, R) >droYak2.chr3L 20703867 97 - 24197627 GGGUUUUUCCGUCAGUUUUGGCUUUUUAAAGCCAAGACUUGUCAGGUGAA-UAGUAAUCUUUG--GAAA-CAG-----AAAUACUUAACCUAAUUAUUAUUUAACU (((....)))(.((((((((((((....)))))))))).)).)((((...-.((((...((((--....-)))-----)..))))..))))............... ( -27.80, z-score = -3.79, R) >droSec1.super_0 700784 101 + 21120651 GGGUUUUUCCGUUAGUUUUGGCUUU--AAAGCUAAGACUUGACUAGUAAACUAGAUCUAGUAG--AAAAAAUAUUAAUGAAAACAU-UACUCAUUAUCAUUUAACU (((((((((..((((((((((((..--..)))))))))..((((((....)))).)))))..)--)))))....(((((....)))-))))).............. ( -21.10, z-score = -2.19, R) >droSim1.chr3L 7865171 96 + 22553184 GGGUUUUUCCGUUAGUUUUGGCUUUU-AAAGCUAGGACUUGAUUAGUAAACUAGUAGUA-------AAAUAAAUUGAUUAAAACA--GCCUCAUUAUCAUUUAACU ((((((((..((((((((....((((-(..(((((.(((.....)))...)))))..))-------))).))))))))..))).)--))))............... ( -17.00, z-score = -0.52, R) >consensus GGGUUUUUCCGUUAGUUUUGGCUUUU_AAAGCUAAGACUUGACUAGUAAACUAGUAAUAGUAG__AAAAAAAAUUAAUAAAAACAUAGCCUCAUUAUCAUUUAACU (((....)))((((((((((((((....)))))))))).))))............................................................... (-11.90 = -11.98 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:11:31 2011