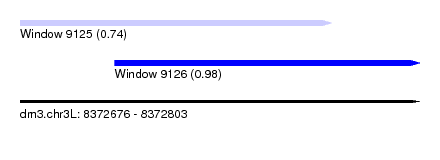
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,372,676 – 8,372,803 |
| Length | 127 |
| Max. P | 0.976756 |

| Location | 8,372,676 – 8,372,775 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.85 |
| Shannon entropy | 0.42225 |
| G+C content | 0.32003 |
| Mean single sequence MFE | -20.41 |
| Consensus MFE | -9.72 |
| Energy contribution | -10.17 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.739529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
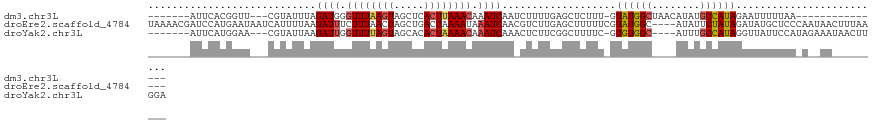
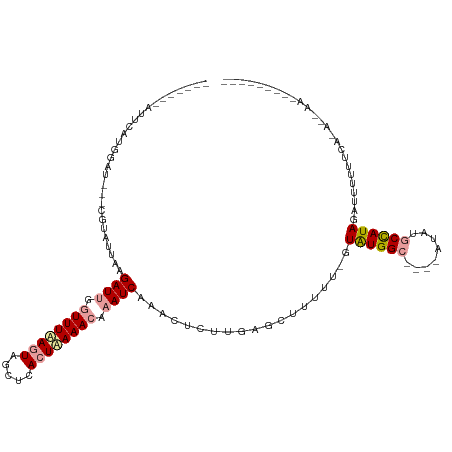
>dm3.chr3L 8372676 99 + 24543557 UUCGUACUUCACAGCUGAAAAUUAAGCGUUA-----------------UUCACGGUU---CGUAUUUAGAUGGGUUUAAGUAGCUCACUUAAACAAAUCAAUCUUUUGAGCUCUUU-GUA ..........((((((........)))....-----------------.....((((---((......(((..((((((((.....))))))))..))).......))))))...)-)). ( -18.22, z-score = -0.80, R) >droEre2.scaffold_4784 20103849 120 - 25762168 UCAGUAAUUCCCAGCUAAAAAUUACGAGUUAAAGAAAUCUUAAAACGAUCCAUGAAUAAUCAUUUUAAGAUUUCUUUAACUAGCUGACUAAAAUAAAUCAACGUCUUGAGCUUUUUCGUA ...........(((((..........(((((((((((((((((((.(((.........))).))))))))))))))))))))))))((.((((....((((....))))...)))).)). ( -28.60, z-score = -4.95, R) >droYak2.chr3L 20671848 99 - 24197627 UUCGUACUUCUCAGCUAAAAAUUAAGUGUUA-----------------UUCAUGGAA---CGUAUUAAGAUUGGUUUUAGUAGCACACUAAAACAAAUCAAACUCUUCGGCUUUUC-GUG ...............................-----------------..(((((((---........((((.((((((((.....)))))))).)))).....(....)..))))-))) ( -14.40, z-score = -0.27, R) >consensus UUCGUACUUCACAGCUAAAAAUUAAGAGUUA_________________UUCAUGGAU___CGUAUUAAGAUUGGUUUAAGUAGCUCACUAAAACAAAUCAAACUCUUGAGCUUUUU_GUA ............((((...................................(((......))).....((((.((((((((.....)))))))).)))).........))))........ ( -9.72 = -10.17 + 0.45)

| Location | 8,372,706 – 8,372,803 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 123 |
| Reading direction | forward |
| Mean pairwise identity | 58.97 |
| Shannon entropy | 0.55441 |
| G+C content | 0.31864 |
| Mean single sequence MFE | -21.54 |
| Consensus MFE | -11.00 |
| Energy contribution | -11.57 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.976756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8372706 97 + 24543557 -------AUUCACGGUU---CGUAUUUAGAUGGGUUUAAGUAGCUCACUUAAACAAAUCAAUCUUUUGAGCUCUUU-GUAUGGCUAACAUAUGCCAUAGAAUUUUUAA--------------- -------((((..((((---((......(((..((((((((.....))))))))..))).......))))))....-.((((((........))))))))))......--------------- ( -23.72, z-score = -3.10, R) >droEre2.scaffold_4784 20103889 116 - 25762168 UAAAACGAUCCAUGAAUAAUCAUUUUAAGAUUUCUUUAACUAGCUGACUAAAAUAAAUCAACGUCUUGAGCUUUUUCGUAUGGC----AUAUUCUAUAGAUAUGCUCCCAAUAACUUUAA--- ....((((...((((....))))((((((((...((((..(((....)))...)))).....)))))))).....))))..(((----(((((.....))))))))..............--- ( -13.70, z-score = -0.04, R) >droYak2.chr3L 20671878 108 - 24197627 -------AUUCAUGGAA---CGUAUUAAGAUUGGUUUUAGUAGCACACUAAAACAAAUCAAACUCUUCGGCUUUUC-GUGUGGC----AUUUGCCAUAGGUUAUUCCAUAGAAAUAACUUGGA -------.(((((((((---........((((.((((((((.....)))))))).))))................(-.((((((----....)))))).)...)))))).))).......... ( -27.20, z-score = -2.68, R) >consensus _______AUUCAUGGAU___CGUAUUAAGAUUGGUUUAAGUAGCUCACUAAAACAAAUCAAACUCUUGAGCUUUUU_GUAUGGC____AUAUGCCAUAGAUUUUUUCA_A__AA_________ ...........(((......))).....((((.((((((((.....)))))))).))))...................((((((........))))))......................... (-11.00 = -11.57 + 0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:11:28 2011