| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,356,566 – 8,356,689 |
| Length | 123 |
| Max. P | 0.764612 |

| Location | 8,356,566 – 8,356,658 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 69.47 |
| Shannon entropy | 0.58842 |
| G+C content | 0.43513 |
| Mean single sequence MFE | -22.51 |
| Consensus MFE | -10.84 |
| Energy contribution | -11.58 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.764612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
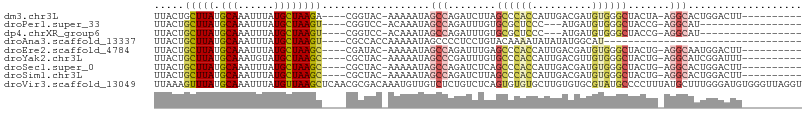
>dm3.chr3L 8356566 92 + 24543557 UUACUGCUUAUGCAAAUUUAUGCUAAGA----CGGUAC-AAAAAUAGCCAGAUCUUAGCCCACCAUUGACGAUGUGGGCUACUA-AGGCACUGGACUU---------- .(((((((((.(((......))))))).----))))).-......((((((.(((((((((((.(((...)))))))))))...-)))..)))).)).---------- ( -27.50, z-score = -3.22, R) >droPer1.super_33 854745 82 + 967471 UUACUGCUUAUGCAAAUUUAUGCUAAGU----CGGUCC-ACAAAUAGCCAGAUUUGUGCGCUCCC---AUGAUGUGGGCUACCG-AGGCAU----------------- ....(((....))).....(((((...(----((((.(-((((((......)))))))....(((---((...)))))..))))-))))))----------------- ( -21.60, z-score = -0.47, R) >dp4.chrXR_group6 12830327 82 + 13314419 UUACUGCUUAUGCAAAUUUAUGCUAAGU----CGGUCC-ACAAAUAGCCAGAUUUGUGCGCUCCC---AUGAUGUGGGCUACCG-AGGCAU----------------- ....(((....))).....(((((...(----((((.(-((((((......)))))))....(((---((...)))))..))))-))))))----------------- ( -21.60, z-score = -0.47, R) >droAna3.scaffold_13337 15857177 71 - 23293914 UUACUGCUUAUGCAAAUUUAUGCUAAGU----CGCCACCAAAAAUAGCCCCUCCUGUACAAAAUAUAUAUGGCAU--------------------------------- .....(((((.(((......))))))))----.((((................................))))..--------------------------------- ( -8.45, z-score = 0.12, R) >droEre2.scaffold_4784 20087911 92 - 25762168 UUACUGCUUAUGCAAAUUUAUGCUAAGC----CGAUAC-AAAAAUAGCCAGAUUUGAGCCCACCAUUGACGAUGUGGGCUACUG-AGGCAAUGGACUU---------- ...(.(((((.(((......))))))))----.)....-.......(((.......(((((((.(((...))))))))))....-.))).........---------- ( -23.80, z-score = -2.11, R) >droYak2.chr3L 20655630 92 - 24197627 UUACUGCUUAUGCAAAUGUAUGCUAAGC----CGCUAC-AAAAAUAGCCCGAUUUGUGCCCACCAUUGACGUUGUGGGCUACUG-AGGCAUCGGAUUU---------- .....(((((.(((......))))))))----.((((.-.....))))(((((..((((((((.((....)).)))))).))..-....)))))....---------- ( -25.70, z-score = -1.30, R) >droSec1.super_0 652914 92 + 21120651 UUACUGCUUAUGCAAAUUUAUGCUAAGC----CGCUAC-AAAAAUAGCCAGAUCUCAGCCCACCAUUGACGAUGUGGGCUACUG-AGGCACUGGACUU---------- .....(((((.(((......))))))))----......-......((((((.(((((((((((.(((...)))))))))...))-)))..)))).)).---------- ( -26.50, z-score = -2.30, R) >droSim1.chr3L 7816778 92 + 22553184 UUACUGCUUAUGCAAAUUUAUGCUAAGC----CGCUAC-AAAAAUAGCCAGAUCUUAGCCCACCAUUGACGAUGUGGGCUACUG-AGGCACUGGACUU---------- .....(((((.(((......))))))))----......-......((((((.((.((((((((.(((...)))))))))))..)-)....)))).)).---------- ( -25.80, z-score = -2.23, R) >droVir3.scaffold_13049 4763141 108 + 25233164 UUAAAGUUUAUGCAAAUUUAUGUUAAGCUCAACGCGACAAAUGUUGUCUCUGUCUCAGUGUGUGCUUGUGUGCGUAUGCCCCUUUAUGCUUUGGGAUGUGGGUUAGGU ....((((((.(((......)))))))))....(((((....))))).((((.((((((((((((......))))))))(((..........)))...)))).)))). ( -21.60, z-score = 0.99, R) >consensus UUACUGCUUAUGCAAAUUUAUGCUAAGC____CGCUAC_AAAAAUAGCCAGAUCUGAGCCCACCAUUGACGAUGUGGGCUACUG_AGGCAUUGGACUU__________ .....(((((.(((......))))))))..................(((........((((((..........)))))).......)))................... (-10.84 = -11.58 + 0.75)


| Location | 8,356,599 – 8,356,689 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 93.67 |
| Shannon entropy | 0.11512 |
| G+C content | 0.48667 |
| Mean single sequence MFE | -27.54 |
| Consensus MFE | -25.14 |
| Energy contribution | -25.18 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.726872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8356599 90 + 24543557 CAAAAAUAGCCAGAUCUUAGCCCACCAUUGACGAUGUGGGCUACUAAGGCACUGGACUUGGGUUCGUCUUCGUCUUAUGCGGCAUUAUUU ........(((......((((((((.(((...))))))))))).....(((..((((..(((....)))..))))..))))))....... ( -27.80, z-score = -2.18, R) >droEre2.scaffold_4784 20087944 90 - 25762168 CAAAAAUAGCCAGAUUUGAGCCCACCAUUGACGAUGUGGGCUACUGAGGCAAUGGACUUGGGUUCGUCUUCGUCUUAUGCGGCAUCAUUU ........(((.......(((((((.(((...))))))))))..((((((...((((........))))..))))))...)))....... ( -26.30, z-score = -1.08, R) >droYak2.chr3L 20655663 90 - 24197627 CAAAAAUAGCCCGAUUUGUGCCCACCAUUGACGUUGUGGGCUACUGAGGCAUCGGAUUUGGGUUCGUCUGCGUCUUUUGCGGCAUCAUUU .......(((((((.....((((((.((....)).))))))..((((....))))..))))))).(.(((((.....))))))....... ( -26.60, z-score = -0.55, R) >droSec1.super_0 652947 90 + 21120651 CAAAAAUAGCCAGAUCUCAGCCCACCAUUGACGAUGUGGGCUACUGAGGCACUGGACUUGGGUUCGUCUUCGUCUUAUGCGGCAUCAUUU ........(((....((((((((((.(((...)))))))))...))))(((..((((..(((....)))..))))..))))))....... ( -28.50, z-score = -1.71, R) >droSim1.chr3L 7816811 90 + 22553184 CAAAAAUAGCCAGAUCUUAGCCCACCAUUGACGAUGUGGGCUACUGAGGCACUGGACUUGGGUUCGUCUUCGUCUUAUGCGGCAUCAUUU ........(((...((.((((((((.(((...)))))))))))..)).(((..((((..(((....)))..))))..))))))....... ( -28.50, z-score = -1.89, R) >consensus CAAAAAUAGCCAGAUCUGAGCCCACCAUUGACGAUGUGGGCUACUGAGGCACUGGACUUGGGUUCGUCUUCGUCUUAUGCGGCAUCAUUU ........(((.......(((((((..........)))))))......(((..((((..(((....)))..))))..))))))....... (-25.14 = -25.18 + 0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:11:24 2011