| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,270,583 – 8,270,683 |
| Length | 100 |
| Max. P | 0.611632 |

| Location | 8,270,583 – 8,270,683 |
|---|---|
| Length | 100 |
| Sequences | 15 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 79.31 |
| Shannon entropy | 0.48678 |
| G+C content | 0.52173 |
| Mean single sequence MFE | -34.72 |
| Consensus MFE | -14.37 |
| Energy contribution | -14.00 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.611632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
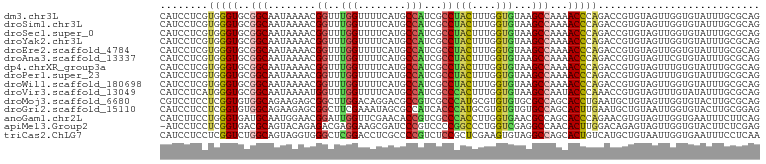
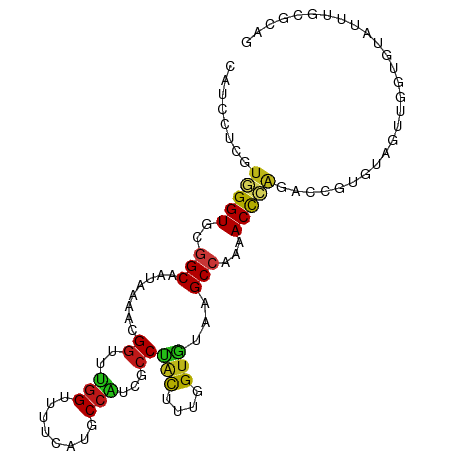
>dm3.chr3L 8270583 100 - 24543557 CAUCCUCGUGGGUGCGGCAAUAAAACGGUUUGGUUUUCAUGCCAUCGCCUACUUUGGUGUAAGCCAAAACCCAGACCGUGUAGUUGGUGUAUUUGCGCAG ....((((..((((((.((((...(((((((((((((...((...((((......))))...)).)))).)))))))))...)))).))))))..)).)) ( -38.20, z-score = -3.32, R) >droSim1.chr3L 7729806 100 - 22553184 CAUCCUCGUGGGUGCGGCAAUAAAACGGUUUGGUUUUCAUGCCAUCGCCUACUUUGGUGUAAGCCAAAACCCAGACCGUGUAGUUGGUGUAUUUGCGCAG ....((((..((((((.((((...(((((((((((((...((...((((......))))...)).)))).)))))))))...)))).))))))..)).)) ( -38.20, z-score = -3.32, R) >droSec1.super_0 551373 100 - 21120651 CAUCCUCGUGGGUGCGGCAAUAAAACGGUUUGGUUUUCAUGCCAUCGCCUACUUUGGUGUAAGCCAAAACCCAGACCGUGUAGUUGGUGUAUUUGCGCAG ....((((..((((((.((((...(((((((((((((...((...((((......))))...)).)))).)))))))))...)))).))))))..)).)) ( -38.20, z-score = -3.32, R) >droYak2.chr3L 20568924 100 + 24197627 CAUCCUCGUGGGUGCGGCAAUAAAACGGUUUGGUUUUCAUGCCAUCGCCUACUUUGGUGUAAGCCAAAACCCAGACCGUGUAGUUGGUGUAUUUGCGCAG ....((((..((((((.((((...(((((((((((((...((...((((......))))...)).)))).)))))))))...)))).))))))..)).)) ( -38.20, z-score = -3.32, R) >droEre2.scaffold_4784 19995765 100 + 25762168 CAUCCUCGUGGGUGCGGCAAUAAAACGGUUUGGUUUUCAUGCCAUCGCCUACUUUGGUGUAAGCCAAAACCCAGACCGUGUAGUUGGUGUAUUUGCGCAG ....((((..((((((.((((...(((((((((((((...((...((((......))))...)).)))).)))))))))...)))).))))))..)).)) ( -38.20, z-score = -3.32, R) >droAna3.scaffold_13337 3272208 100 - 23293914 CAUCCUCGUGGGUGCGGCAAUAAAACGGUUUGGUUUUCAUGCCAUCGCCUACUUUGGUGUAAGCCAAAACCCAGACCGUGUAGUUCGUGUAUUUGCGCAG ....((((..((((((..(((...(((((((((((((...((...((((......))))...)).)))).)))))))))...)))..))))))..)).)) ( -32.40, z-score = -1.99, R) >dp4.chrXR_group3a 1424785 100 + 1468910 CAUCCUCGUGGGUGCGGCAAUAAAACGGUUUGGUUUUCAUGCCAUCGCCUACUUUGGUGUAAGCCAAAACCCAGACCGUGUAGUUUGUGUAUUUGCGCAG ....((((..((((((.(((....(((((((((((((...((...((((......))))...)).)))).))))))))).....)))))))))..)).)) ( -34.30, z-score = -2.53, R) >droPer1.super_23 1617724 100 + 1662726 CAUCCUCGUGGGUGCGGCAAUAAAACGGUUUGGUUUUCAUGCCAUCGCCUACUUUGGUGUAAGCCAAAACCCAGACCGUGUAGUUUGUGUAUUUGCGCAG ....((((..((((((.(((....(((((((((((((...((...((((......))))...)).)))).))))))))).....)))))))))..)).)) ( -34.30, z-score = -2.53, R) >droWil1.scaffold_180698 2228937 100 - 11422946 CAUCCUCGUGGGUGCGGCAAUAAAACGGUUUGGUUUUCAUGCCAUCGCCUACUUUGGUGUAAGCCAAAACCCAGACCGUGUAGUUGGUGUAUUUGCGCAG ....((((..((((((.((((...(((((((((((((...((...((((......))))...)).)))).)))))))))...)))).))))))..)).)) ( -38.20, z-score = -3.32, R) >droVir3.scaffold_13049 22703825 100 + 25233164 CAUCCUCAUGGGUGCGGCAAUAAAAUGGUUUGGUUUUCAUGCCAUCGCCCACUUUGGUGUAAGCCAAUACCCAAACCGUGUAGUUUGUAUAUUUGCGCAG ...((....))(((((((((....(((((((((.......((...((((......))))...))......))))))))).....)))).....))))).. ( -26.42, z-score = -0.48, R) >droMoj3.scaffold_6680 3789531 100 + 24764193 CGUCCUCCUCGGUGUGGCAGAAGAGCGGCUUGGACAGGACGCCGUCGCCCAUGCGUGUGUGCGCCAGCACCUGAAUGCUGUAGUUGGUGUACUUGCGCAG ((((((..((((.((.((......)).))))))..))))))..........((((((.((((((((((..(........)..)))))))))).)))))). ( -38.30, z-score = 0.11, R) >droGri2.scaffold_15110 11756129 100 + 24565398 CAUCCUCCUCGGUGUGGCAGAAGAGCGGCUUCGAAAUAGCGCCAUCACCCAUGCGUGUGUGUGCCAGCACUUGAAUGCUGUAAUUGGUGUACUUGCGGAG ....((((..((((((((.((((.....))))(......))))).))))...(((...(((..(((((((......).)))...)))..))).))))))) ( -31.20, z-score = 0.05, R) >anoGam1.chr2L 41969269 100 - 48795086 CAUCUUCCUGGGUGAUGCAAUGGAACGGAUUGGUUCGAACACCGUCGCCCACCUUGGUGAACGCCAGCACCCAGAACGUGUAGUUGGUGAAUUUCUUCAG ((((....(((((((((...(.((((......)))).)....)))))))))....))))..(((((((((.(.....).)).)))))))........... ( -30.80, z-score = -0.59, R) >apiMel3.Group2 9905177 99 + 14465785 -AUCCUCCUCGGUGACGCAGUACAGAGACGAGGAAGCGAUCCCGUCCCCGGCCCUGGUCGAGGCCAACACUUGGACAGAGUAGUUGGUGUACUUCUCGAG -......(((((.((...((((((..((((.(((.....)))))))((((((....)))).))(((((((((.....)))).)))))))))))))))))) ( -34.10, z-score = -0.38, R) >triCas2.ChLG7 14277400 100 + 17478683 CAUCCUCCUCGGUCUGGCAGUAGGUGGGCUCGGACCUCGCCCCGUCUCCGCUCGAAGUGUAGGCCAGCACUGUCAUGCUGUAAUUGGUGAAUUUCCUCAA ((((.....((((.(((((((.((((((......)).))))..((((.(((.....))).))))....))))))).)))).....))))........... ( -29.80, z-score = 0.17, R) >consensus CAUCCUCGUGGGUGCGGCAAUAAAACGGUUUGGUUUUCAUGCCAUCGCCUACUUUGGUGUAAGCCAAAACCCAGACCGUGUAGUUGGUGUAUUUGCGCAG ........(((((..(((........((..(((........)))...))(((....)))...)))...)))))........................... (-14.37 = -14.00 + -0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:11:16 2011