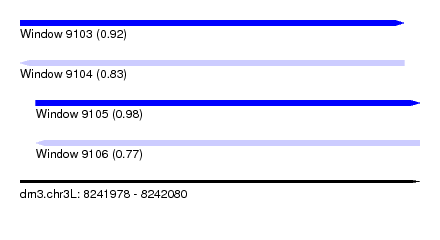
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,241,978 – 8,242,080 |
| Length | 102 |
| Max. P | 0.979611 |

| Location | 8,241,978 – 8,242,076 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.77 |
| Shannon entropy | 0.39481 |
| G+C content | 0.38225 |
| Mean single sequence MFE | -21.35 |
| Consensus MFE | -15.57 |
| Energy contribution | -15.85 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.919885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8241978 98 + 24543557 AAGCAUUAGUGCUCAUAAAAAAUGGAUUUACACAGCCUUUUUAAGGCCAUCCUGAUCCUAUAUGUCAUAAGAGGUGAGUAUUUCUCUUUUUUUCGUUU--------- (((....(((((((((.......(((((......((((.....))))......)))))......((....)).)))))))))...)))..........--------- ( -18.30, z-score = -0.76, R) >droSim1.chr3L 7700723 97 + 22553184 AAGCAUUAGUGCUCAUAAAAAAUGGAUUUACACAGCCUUUUUAAGGCCAUCCUGAUCCUAUAUGUCAUAAGAGGUGAGUAUUUCCCGUUU-UUCGUUU--------- ((((...(((((((((.......(((((......((((.....))))......)))))......((....)).)))))))))....))))-.......--------- ( -19.30, z-score = -1.21, R) >droSec1.super_0 522687 97 + 21120651 AAGCAUUAGUGCUCAUAAAAAAUGGAUUUACACAGCCUUUUUAAGGCCAUCCUGAUCCUAUAUGUCAUAAGUGGUGAGUAUUUCCCUUUU-UCCGUUU--------- (((....(((((((((.......(((((......((((.....))))......))))).......((....)))))))))))...)))..-.......--------- ( -18.00, z-score = -0.86, R) >droYak2.chr3L 20537349 98 - 24197627 AAGCAUUAGUGCUCAUAAAAAAUGGAUUUACACAGCCUUUUUAAGGCCAUCCUGAUCCUAUAUGUCAUAAGAGGUGAGUAUUUCUCUUUUAUUUGUUU--------- (((....(((((((((.......(((((......((((.....))))......)))))......((....)).)))))))))...)))..........--------- ( -18.30, z-score = -0.55, R) >droEre2.scaffold_4784 19966115 97 - 25762168 AAGCAUUAGUGCUCAUAAAAAAUGGAUUUACACAGCCUUUUUAAGGCCAUCCUGAUCCUAUAUGUCAUCAGAGGUGAGUACUUCUCUUUU-UUCGCUU--------- .((((....))))...((((((.(((..(((...((((.....))))(((((((((.(.....)..))))).)))).)))..))).))))-)).....--------- ( -20.80, z-score = -1.33, R) >droAna3.scaffold_13337 3242278 98 + 23293914 AAGCAUUAGUGCUCAUAAAAAAUGGAUUUACACAGCCUUUUUAAGGCCAUCCUGAUCCUGUAUGUCAUACGAGGUGAGUAUUUCCAGCCAGUUUUCCU--------- ..((...(((((((((.......(((((......((((.....))))......)))))(((((...)))))..)))))))))....))..........--------- ( -21.50, z-score = -1.19, R) >dp4.chrXR_group3a 1394748 101 - 1468910 AAGCAUUAGUGCUCAUAAAAAAUGGAUUUACAUAGCCUUUUUAAGGCCGUCCUGAUCCUGUAUGUCAUACGAGGUGAGUAUUAGCCCGC------CUCCUCCCUUCC .((((....))))((((......(((((...(..((((.....))))..)...)))))..))))......((((((.((....)).)))------)))......... ( -23.80, z-score = -1.85, R) >droPer1.super_23 1587284 107 - 1662726 AAGCAUUAGUGCUCAUAAAAAAUGGAUUUACAUAGCCUUUUUAAGGCCGUCCUGAUCCUGUAUGUCAUACGAGGUGAGUAUUAGCCCGCAGAUACCUCCUCCCUUCC .((((....))))((((......(((((...(..((((.....))))..)...)))))..))))......((((.(.(((((.(....).)))))).))))...... ( -24.20, z-score = -1.45, R) >droWil1.scaffold_180698 2191142 83 + 11422946 AAGCAUUAGUGCUCAUAAAAAAUGGAUUUUUACAGCCUUUUUAAGGCUGUCCUGAUCCUGUAUGUCAUACAAGGUGAGUAACC------------------------ .........(((((((.......(((((...(((((((.....)))))))...)))))(((((...)))))..)))))))...------------------------ ( -25.40, z-score = -3.59, R) >droVir3.scaffold_13049 22667330 79 - 25233164 AAGCAUUAGUGCUCAUAAAAAAUGGAUUUACACAGCCUUUUUAAGGCUGUCCUGAUCCUGUACAUCAUACGAGGUGAGU---------------------------- ..........((((((.......(((((...(((((((.....)))))))...)))))((((.....))))..))))))---------------------------- ( -22.50, z-score = -2.66, R) >droMoj3.scaffold_6654 1320052 80 + 2564135 AAGCAUUAGUGCUCAUAAAAAAUGGAUUUACACGGCCUUUUUAAGGCUGUCCUGAUCCUGUACAUCAUACGAGGUGAGUA--------------------------- .........(((((((.......(((((...(((((((.....)))))))...)))))((((.....))))..)))))))--------------------------- ( -22.80, z-score = -2.43, R) >consensus AAGCAUUAGUGCUCAUAAAAAAUGGAUUUACACAGCCUUUUUAAGGCCAUCCUGAUCCUGUAUGUCAUACGAGGUGAGUAUUUCCCUUUU_UU_GCUU_________ .........(((((((.......(((((......((((.....))))......)))))......((....)).)))))))........................... (-15.57 = -15.85 + 0.27)

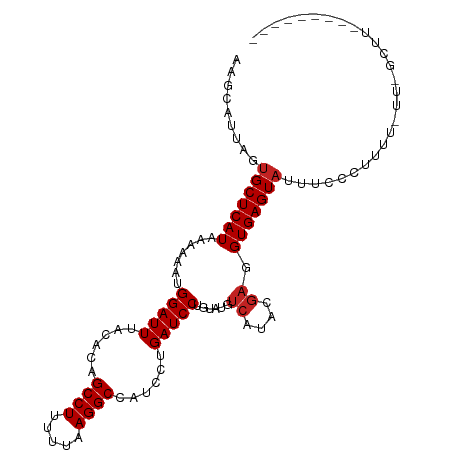
| Location | 8,241,978 – 8,242,076 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.77 |
| Shannon entropy | 0.39481 |
| G+C content | 0.38225 |
| Mean single sequence MFE | -20.72 |
| Consensus MFE | -16.52 |
| Energy contribution | -16.60 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.829794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8241978 98 - 24543557 ---------AAACGAAAAAAAGAGAAAUACUCACCUCUUAUGACAUAUAGGAUCAGGAUGGCCUUAAAAAGGCUGUGUAAAUCCAUUUUUUAUGAGCACUAAUGCUU ---------....(((((((((((..........)))))..........((((..(.(((((((.....))))))).)..)))).))))))..(((((....))))) ( -21.00, z-score = -2.02, R) >droSim1.chr3L 7700723 97 - 22553184 ---------AAACGAA-AAACGGGAAAUACUCACCUCUUAUGACAUAUAGGAUCAGGAUGGCCUUAAAAAGGCUGUGUAAAUCCAUUUUUUAUGAGCACUAAUGCUU ---------....(((-(((..(((..(((...(((((((((...)))))))...))(((((((.....))))))))))..))).))))))..(((((....))))) ( -19.90, z-score = -1.33, R) >droSec1.super_0 522687 97 - 21120651 ---------AAACGGA-AAAAGGGAAAUACUCACCACUUAUGACAUAUAGGAUCAGGAUGGCCUUAAAAAGGCUGUGUAAAUCCAUUUUUUAUGAGCACUAAUGCUU ---------.....((-((((((((.....)).))..............((((..(.(((((((.....))))))).)..)))).))))))..(((((....))))) ( -20.10, z-score = -1.15, R) >droYak2.chr3L 20537349 98 + 24197627 ---------AAACAAAUAAAAGAGAAAUACUCACCUCUUAUGACAUAUAGGAUCAGGAUGGCCUUAAAAAGGCUGUGUAAAUCCAUUUUUUAUGAGCACUAAUGCUU ---------......(((((((((.....))).................((((..(.(((((((.....))))))).)..))))...))))))(((((....))))) ( -20.00, z-score = -1.63, R) >droEre2.scaffold_4784 19966115 97 + 25762168 ---------AAGCGAA-AAAAGAGAAGUACUCACCUCUGAUGACAUAUAGGAUCAGGAUGGCCUUAAAAAGGCUGUGUAAAUCCAUUUUUUAUGAGCACUAAUGCUU ---------....(((-(((((((..........))))...........((((..(.(((((((.....))))))).)..)))).))))))..(((((....))))) ( -20.10, z-score = -0.70, R) >droAna3.scaffold_13337 3242278 98 - 23293914 ---------AGGAAAACUGGCUGGAAAUACUCACCUCGUAUGACAUACAGGAUCAGGAUGGCCUUAAAAAGGCUGUGUAAAUCCAUUUUUUAUGAGCACUAAUGCUU ---------.(((...(((..((...((((.......))))..))..))).....(.(((((((.....))))))).)...))).........(((((....))))) ( -20.90, z-score = -0.34, R) >dp4.chrXR_group3a 1394748 101 + 1468910 GGAAGGGAGGAG------GCGGGCUAAUACUCACCUCGUAUGACAUACAGGAUCAGGACGGCCUUAAAAAGGCUAUGUAAAUCCAUUUUUUAUGAGCACUAAUGCUU .........(((------(.(((......))).))))((((...)))).((((..(.(..((((.....))))..).)..)))).........(((((....))))) ( -23.90, z-score = -0.52, R) >droPer1.super_23 1587284 107 + 1662726 GGAAGGGAGGAGGUAUCUGCGGGCUAAUACUCACCUCGUAUGACAUACAGGAUCAGGACGGCCUUAAAAAGGCUAUGUAAAUCCAUUUUUUAUGAGCACUAAUGCUU ......((((.(((((..(....)..)))))..))))((((...)))).((((..(.(..((((.....))))..).)..)))).........(((((....))))) ( -24.10, z-score = 0.11, R) >droWil1.scaffold_180698 2191142 83 - 11422946 ------------------------GGUUACUCACCUUGUAUGACAUACAGGAUCAGGACAGCCUUAAAAAGGCUGUAAAAAUCCAUUUUUUAUGAGCACUAAUGCUU ------------------------(((.....))).(((((...)))))((((....(((((((.....)))))))....)))).........(((((....))))) ( -21.30, z-score = -2.51, R) >droVir3.scaffold_13049 22667330 79 + 25233164 ----------------------------ACUCACCUCGUAUGAUGUACAGGAUCAGGACAGCCUUAAAAAGGCUGUGUAAAUCCAUUUUUUAUGAGCACUAAUGCUU ----------------------------.((((....((((...)))).((((..(.(((((((.....))))))).)..))))........))))........... ( -20.60, z-score = -2.23, R) >droMoj3.scaffold_6654 1320052 80 - 2564135 ---------------------------UACUCACCUCGUAUGAUGUACAGGAUCAGGACAGCCUUAAAAAGGCCGUGUAAAUCCAUUUUUUAUGAGCACUAAUGCUU ---------------------------..((((....((((...)))).((((..(.((.((((.....)))).)).)..))))........))))........... ( -16.00, z-score = -0.71, R) >consensus _________AAAC_AA_AAAAGGGAAAUACUCACCUCGUAUGACAUACAGGAUCAGGAUGGCCUUAAAAAGGCUGUGUAAAUCCAUUUUUUAUGAGCACUAAUGCUU .................................................((((..(.(((((((.....))))))).)..)))).........(((((....))))) (-16.52 = -16.60 + 0.08)

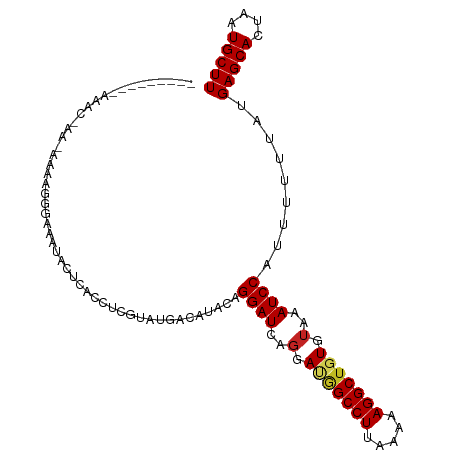
| Location | 8,241,982 – 8,242,080 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 79.42 |
| Shannon entropy | 0.42142 |
| G+C content | 0.38131 |
| Mean single sequence MFE | -20.97 |
| Consensus MFE | -15.57 |
| Energy contribution | -15.85 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8241982 98 + 24543557 AUUAGUGCUCAUAAAAAAUGGAUUUACACAGCCUUUUUAAGGCCAUCCUGAUCCUAUAUGUCAUAAGAGGUGAGUAUUUCUCUUUUUUU--CGUUUCCAU----- ...(((((((((.......(((((......((((.....))))......)))))......((....)).)))))))))...........--.........----- ( -18.20, z-score = -1.07, R) >droSim1.chr3L 7700727 97 + 22553184 AUUAGUGCUCAUAAAAAAUGGAUUUACACAGCCUUUUUAAGGCCAUCCUGAUCCUAUAUGUCAUAAGAGGUGAGUAUUUCCCGUUUUU---CGUUUCCAU----- ...(((((((((.......(((((......((((.....))))......)))))......((....)).)))))))))..........---.........----- ( -18.20, z-score = -1.15, R) >droSec1.super_0 522691 97 + 21120651 AUUAGUGCUCAUAAAAAAUGGAUUUACACAGCCUUUUUAAGGCCAUCCUGAUCCUAUAUGUCAUAAGUGGUGAGUAUUUCCCUUUUUC---CGUUUCCAU----- ...(((((((((.......(((((......((((.....))))......))))).......((....)))))))))))..........---.........----- ( -17.90, z-score = -1.24, R) >droYak2.chr3L 20537353 98 - 24197627 AUUAGUGCUCAUAAAAAAUGGAUUUACACAGCCUUUUUAAGGCCAUCCUGAUCCUAUAUGUCAUAAGAGGUGAGUAUUUCUCUUUUAUU--UGUUUCUAU----- ....(((..((((......(((((......((((.....))))......)))))..)))).))).(((((..((((.........))))--..)))))..----- ( -18.40, z-score = -0.87, R) >droEre2.scaffold_4784 19966119 97 - 25762168 AUUAGUGCUCAUAAAAAAUGGAUUUACACAGCCUUUUUAAGGCCAUCCUGAUCCUAUAUGUCAUCAGAGGUGAGUACUUCUCUUUUUU---CGCUUCCAU----- ...(((((((((.......(((((......((((.....))))......)))))......((....)).)))))))))..........---.........----- ( -20.50, z-score = -1.66, R) >droAna3.scaffold_13337 3242282 98 + 23293914 AUUAGUGCUCAUAAAAAAUGGAUUUACACAGCCUUUUUAAGGCCAUCCUGAUCCUGUAUGUCAUACGAGGUGAGUAUUUCCAGCCAGUU--UUCCUCCAU----- ...(((((((((.......(((((......((((.....))))......)))))(((((...)))))..)))))))))...........--.........----- ( -19.60, z-score = -0.89, R) >dp4.chrXR_group3a 1394752 99 - 1468910 AUUAGUGCUCAUAAAAAAUGGAUUUACAUAGCCUUUUUAAGGCCGUCCUGAUCCUGUAUGUCAUACGAGGUGAGUAUUAGCCCGC------CUCCUCCCUUCCCC ....(((..((((......(((((...(..((((.....))))..)...)))))..))))...)))((((((.((....)).)))------)))........... ( -23.40, z-score = -2.26, R) >droPer1.super_23 1587288 105 - 1662726 AUUAGUGCUCAUAAAAAAUGGAUUUACAUAGCCUUUUUAAGGCCGUCCUGAUCCUGUAUGUCAUACGAGGUGAGUAUUAGCCCGCAGAUACCUCCUCCCUUCCCC ....(((..((((......(((((...(..((((.....))))..)...)))))..))))...)))((((.(.(((((.(....).)))))).))))........ ( -23.80, z-score = -1.80, R) >droWil1.scaffold_180698 2191146 79 + 11422946 AUUAGUGCUCAUAAAAAAUGGAUUUUUACAGCCUUUUUAAGGCUGUCCUGAUCCUGUAUGUCAUACAAGGUGAGUAACC-------------------------- .....(((((((.......(((((...(((((((.....)))))))...)))))(((((...)))))..)))))))...-------------------------- ( -25.40, z-score = -4.28, R) >droVir3.scaffold_13049 22667334 75 - 25233164 AUUAGUGCUCAUAAAAAAUGGAUUUACACAGCCUUUUUAAGGCUGUCCUGAUCCUGUACAUCAUACGAGGUGAGU------------------------------ ......((((((.......(((((...(((((((.....)))))))...)))))((((.....))))..))))))------------------------------ ( -22.50, z-score = -3.12, R) >droMoj3.scaffold_6654 1320056 76 + 2564135 AUUAGUGCUCAUAAAAAAUGGAUUUACACGGCCUUUUUAAGGCUGUCCUGAUCCUGUACAUCAUACGAGGUGAGUA----------------------------- .....(((((((.......(((((...(((((((.....)))))))...)))))((((.....))))..)))))))----------------------------- ( -22.80, z-score = -2.95, R) >consensus AUUAGUGCUCAUAAAAAAUGGAUUUACACAGCCUUUUUAAGGCCAUCCUGAUCCUGUAUGUCAUACGAGGUGAGUAUUUCCCUUUU_U___CGCUUCCAU_____ .....(((((((.......(((((......((((.....))))......)))))......((....)).)))))))............................. (-15.57 = -15.85 + 0.27)

| Location | 8,241,982 – 8,242,080 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.42 |
| Shannon entropy | 0.42142 |
| G+C content | 0.38131 |
| Mean single sequence MFE | -19.83 |
| Consensus MFE | -14.63 |
| Energy contribution | -14.72 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.772195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8241982 98 - 24543557 -----AUGGAAACG--AAAAAAAGAGAAAUACUCACCUCUUAUGACAUAUAGGAUCAGGAUGGCCUUAAAAAGGCUGUGUAAAUCCAUUUUUUAUGAGCACUAAU -----.((....))--.......(((.....)))....(((((((...((.((((..(.(((((((.....))))))).)..))))))...)))))))....... ( -20.30, z-score = -1.70, R) >droSim1.chr3L 7700727 97 - 22553184 -----AUGGAAACG---AAAAACGGGAAAUACUCACCUCUUAUGACAUAUAGGAUCAGGAUGGCCUUAAAAAGGCUGUGUAAAUCCAUUUUUUAUGAGCACUAAU -----.((....))---......((((.....)).)).(((((((...((.((((..(.(((((((.....))))))).)..))))))...)))))))....... ( -20.00, z-score = -1.29, R) >droSec1.super_0 522691 97 - 21120651 -----AUGGAAACG---GAAAAAGGGAAAUACUCACCACUUAUGACAUAUAGGAUCAGGAUGGCCUUAAAAAGGCUGUGUAAAUCCAUUUUUUAUGAGCACUAAU -----.((....))---......((((.....)).)).(((((((...((.((((..(.(((((((.....))))))).)..))))))...)))))))....... ( -20.80, z-score = -1.48, R) >droYak2.chr3L 20537353 98 + 24197627 -----AUAGAAACA--AAUAAAAGAGAAAUACUCACCUCUUAUGACAUAUAGGAUCAGGAUGGCCUUAAAAAGGCUGUGUAAAUCCAUUUUUUAUGAGCACUAAU -----.(((.....--.......(((.....)))....(((((((...((.((((..(.(((((((.....))))))).)..))))))...)))))))..))).. ( -19.50, z-score = -1.74, R) >droEre2.scaffold_4784 19966119 97 + 25762168 -----AUGGAAGCG---AAAAAAGAGAAGUACUCACCUCUGAUGACAUAUAGGAUCAGGAUGGCCUUAAAAAGGCUGUGUAAAUCCAUUUUUUAUGAGCACUAAU -----......((.---((((((..((..(((.....((((((..(.....).))))))(((((((.....))))))))))..))..))))))....))...... ( -19.50, z-score = -0.61, R) >droAna3.scaffold_13337 3242282 98 - 23293914 -----AUGGAGGAA--AACUGGCUGGAAAUACUCACCUCGUAUGACAUACAGGAUCAGGAUGGCCUUAAAAAGGCUGUGUAAAUCCAUUUUUUAUGAGCACUAAU -----(((((.((.--..(((..((...((((.......))))..))..)))..)).(.(((((((.....))))))).)...)))))................. ( -19.10, z-score = 0.23, R) >dp4.chrXR_group3a 1394752 99 + 1468910 GGGGAAGGGAGGAG------GCGGGCUAAUACUCACCUCGUAUGACAUACAGGAUCAGGACGGCCUUAAAAAGGCUAUGUAAAUCCAUUUUUUAUGAGCACUAAU ...........(((------(.(((......))).))))((((...)))).((((..(.(..((((.....))))..).)..))))................... ( -20.80, z-score = 0.26, R) >droPer1.super_23 1587288 105 + 1662726 GGGGAAGGGAGGAGGUAUCUGCGGGCUAAUACUCACCUCGUAUGACAUACAGGAUCAGGACGGCCUUAAAAAGGCUAUGUAAAUCCAUUUUUUAUGAGCACUAAU ........((((.(((((..(....)..)))))..))))((((...)))).((((..(.(..((((.....))))..).)..))))................... ( -21.00, z-score = 0.93, R) >droWil1.scaffold_180698 2191146 79 - 11422946 --------------------------GGUUACUCACCUUGUAUGACAUACAGGAUCAGGACAGCCUUAAAAAGGCUGUAAAAAUCCAUUUUUUAUGAGCACUAAU --------------------------(((..((((...(((((...)))))((((....(((((((.....)))))))....))))........)))).)))... ( -20.50, z-score = -2.97, R) >droVir3.scaffold_13049 22667334 75 + 25233164 ------------------------------ACUCACCUCGUAUGAUGUACAGGAUCAGGACAGCCUUAAAAAGGCUGUGUAAAUCCAUUUUUUAUGAGCACUAAU ------------------------------.((((....((((...)))).((((..(.(((((((.....))))))).)..))))........))))....... ( -20.60, z-score = -2.80, R) >droMoj3.scaffold_6654 1320056 76 - 2564135 -----------------------------UACUCACCUCGUAUGAUGUACAGGAUCAGGACAGCCUUAAAAAGGCCGUGUAAAUCCAUUUUUUAUGAGCACUAAU -----------------------------..((((....((((...)))).((((..(.((.((((.....)))).)).)..))))........))))....... ( -16.00, z-score = -1.32, R) >consensus _____AUGGAAACG___A_AAAAGGGAAAUACUCACCUCGUAUGACAUACAGGAUCAGGAUGGCCUUAAAAAGGCUGUGUAAAUCCAUUUUUUAUGAGCACUAAU ...............................((((..((....))......((((..(.(((((((.....))))))).)..))))........))))....... (-14.63 = -14.72 + 0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:11:10 2011